AUCTORES
Globalize your Research
Review Article | DOI: https://doi.org/10.31579/2640-1053/195
1Riggs Pharmaceuticals Department of Pharmacy, University of Karachi
2Assistant Professor, Department of Microbiology, University of Karachi, Pakistan
3Head of department of Pharmacology Fazaia Ruth Pfau Air University, Karachi
4GD Pharmaceutical Inc OPJS University Rajasthan.
5Assistant Professor Department of Pathology, Dow University of Health Sciences Karachi Pakistan.
*Corresponding Author: Rehan Haider, Riggs Pharmaceuticals Department of Pharmacy, University of Karachi.
Citation: Rehan Haider, (2024), Evaluation of cell cycle inhibitors by flow cytometry, J. Cancer Research and Cellular Therapeutics, 8(3); DOI:10.31579/2640-1053/195
Copyright: © 2024, Rehan Haider. this is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Received: 10 April 2024 | Accepted: 18 April 2024 | Published: 25 April 2024
Keywords: cell cycle inhibitors; flow cytometry; DNA staining; cell cycle distribution; cell cycle progression; therapy; high-throughput analysis
The evaluation of cell-cycle inhibitors using flow cytometry is crucial for understanding the mechanism of action of investigational drugs and identifying potential therapeutic targets in cancer treatment. Flow cytometry enables high-throughput analysis of individual cells within a population, providing valuable insights into cell cycle progression and identifying drug-induced changes.
Typically, cells are stained with fluorescent dyes that bind to DNA, allowing for analysis of cell cycle phases based on DNA content. Following treatment with cell cycle inhibitors, alterations in cell cycle progression, such as arrest at specific phases or induction of apoptosis, can be detected by changes in cell population distribution at various time points.
Flow cytometry offers several advantages for the assessment of cell cycle inhibitors, including the ability to detect subtle changes in cell cycle distribution, rapid data acquisition, and compatibility with complementary assays. Additionally, flow cytometry can be combined with other techniques, such as immunostaining or cell sorting, to provide additional mechanistic insights or isolate specific cell populations for further analysis.
However, there are challenges associated with flow cytometry, including the need for consistent protocols to ensure reproducible results, optimization of staining conditions to minimize artifacts, and data analysis for accurate interpretation of complex cell cycle profiles. Despite these challenges, flow cytometry remains an indispensable tool for preclinical and clinical studies aimed at elucidating the mechanisms of action of cell cycle inhibitors and developing novel antitumor therapies.
The regulation of the cell cycle is critical for cellular function and overall tissue homeostasis, and its dysregulation is implicated in various diseases and pathologies. The cell cycle progresses through a series of tightly regulated events involving DNA replication, DNA damage repair, and cell division, orchestrated by a complex interplay of signaling pathways and regulatory proteins. Disruption of these processes can lead to uncontrolled cell proliferation and tumor formation, making the cell cycle an attractive target for therapeutic intervention.
Cell-cycle inhibitors represent a promising class of drugs designed to target cell-cycle progression and halt aberrant cell proliferation, offering potential avenues for the treatment of tumors and various proliferative disorders. Understanding the mechanisms of action of these inhibitors and assessing their efficacy are crucial steps in the development of novel therapeutic strategies.
Flow cytometry has emerged as a powerful technique for evaluating cell-cycle inhibitors, enabling the rapid and quantitative analysis of large numbers of individual cells. By leveraging the characteristic DNA content of cells at different stages of the cell cycle, flow cytometry facilitates the characterization of cell-cycle distribution and the detection of alterations induced by inhibitors.
In this study, we aim to assess the effects of cell-cycle inhibitors using flow cytometry and investigate their impact on long-term progression. Specifically, we will examine the ability of these inhibitors to induce cell-cycle arrest or apoptosis in tumor cells, providing insights into their mechanisms of action and potential therapeutic utility. By utilizing the capabilities of flow cytometry and elucidating the effects of cell-cycle inhibitors on cell-cycle dynamics, this research has the potential to advance our understanding of novel anti-tumor therapies and shed light on cell-cycle requirements in health and disease.
Drug Discovery Process
The primary approach employed by many pharmaceutical companies for drug discovery involves identifying a fundamental biological target that has been sufficiently validated. Typically, biochemical or cellular assays are developed and utilized to screen hits from large compound libraries in the case of new chemical entities (NCEs), as identifying potential lead compounds requires pharmacological activity. These compounds serve as starting points for chemical modifications that influence structure-activity relationships (SARs). The subsequent stage of lead optimization (LO) involves further refining lead compounds to exhibit desired activities in relevant assays, including cellular assays and indications of in vivo efficacy related to pharmacodynamic biomarker expression. The progression of lead compounds is often influenced by potency, selectivity, or efficacy in animal models, as well as pharmacokinetic properties (e.g., oral bioavailability). During this phase, a comprehensive assessment of toxicological findings and rigorous evaluation in preclinical animal models (e.g., xenograft or transgenic animal models) for confirmed therapeutic efficacy is conducted, ultimately leading to the selection of the final candidate for potential clinical trials. In the intricate process of drug discovery, particularly for cell-cycle inhibitors, flow cytometry (FCM) plays a crucial role in two stages. Firstly, in target validation, where demonstration of the involvement of a specific target triggers a change in the cell cycle, and secondly, in the use of cell-based assays to identify compounds that demonstrate that the observed alterations are indeed due to the expected mechanism of action. FCM serves as a valuable readout and a key tool for evaluating the mechanism of action of drugs that impact cell proliferation, as it is rapid, accurate, and amenable to high-throughput analysis. In this context, we provide an overview of the logistical and technical aspects of flow cytometry as a primary tool for drug discovery, emphasizing its utility in assessing the efficacy of novel treatments. Specifically, when identifying the activity of a target or mechanism induced by a compound, a single-parameter DNA analysis is valuable, focusing on the speed and throughput of samples facilitated by automated control. Alternatively, when the mechanism of action of a drug is the focus of the study, a two-parameter analysis such as bromodeoxyuridine (BrdU) or ethyl-deoxy uridine (EdU) incorporation (detected by "click chemistry") along with DNA staining may be employed.
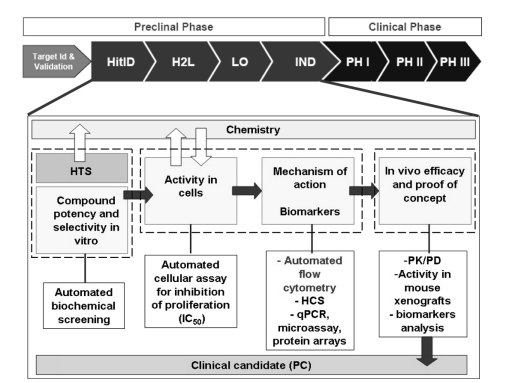
Figure 1. A typical screening approach used for the identification of small molecule cell cycle Inhibitor
2.1 Mono-parametric Analysis of DNA Content
The appraisal of DNA content utilizing glowing dyes that bind stoichiometrically to DNA, such as propidium iodide (PI), allows for the calculation of fundamental DNA content and the dispersion of the container cycle in a population of asynchronously separating containers. Following situations where drugs or siRNA address particular container era stages, alterations in container cycle progress may be discovered. The calculation of DNA content (x-spindle, fluorescence of dye-bound DNA), exhibitivity of the DNA content, and the y-pole interpreting the number of containers is determined utilizing an analytical model (in the way that Modfit) that calculates the distribution of containers in the various stages of the container phase. Proliferating containers progress through three main chapters: G1, S-time, and G2/M. The G2 and M phases include the development of containers by growing their DNA content before dividing into offspring G1 containers. The part of containers accompanying DNA content below the G1 point, frequently referred to as the "Sub G1" part, shows apoptotic and fragmented containers. The incidence of polyploidy in containers, signifying cells accompanying odd DNA content, is lower than in G2/M, primarily on account of the failure in cellular division.
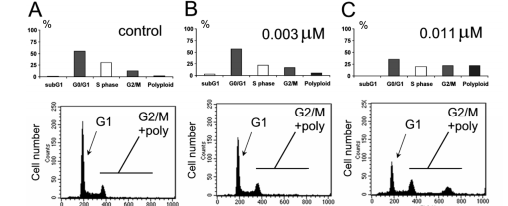
An explanatory model is bestowed in Fig. 2. Human ovarian tumor A2780 cells with a hasty increase were resolved for their DNA content. Various modes of operation in compound situations concede the possibility of being noticed. For instance, doctoring A2780 cells accompanying compounds moving through various stages of the container era may influence unconnected container phase characterizations. For example, treating A2780 containers accompanying compounds moving kinase inhibitors grants permission to lead to various container era sketches, displaying a block before (e.g., CDKs) or following in position or time (like, PLK or Aurora) DNA copy (Table 1 and Fig. 3).
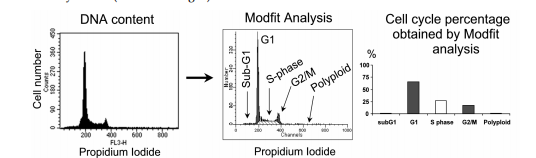
Figure 2. A2780 cells in exponential growth were analyzed for their DNA content and the
percentage of cells in specific stages of the cell cycle percentage was analyzed (X-axis, fluorescence of the dye bound to DNA, representative of the DNA content and y-axis, number of cells to DNA content).
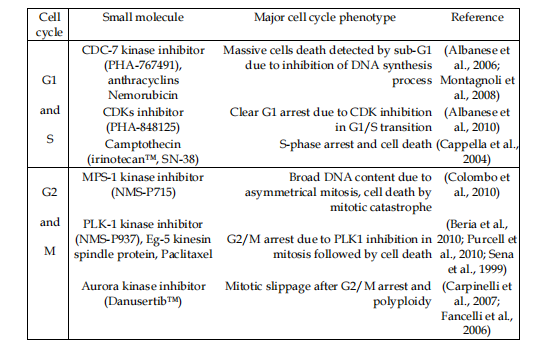
Table 1. Different cellular phenotypes observed after treatment listed as exemplified for small molecule kinase inhibitors targeting cell cycle kinase or cytotoxic agents.
Propidium iodide (Red Fluorescence)
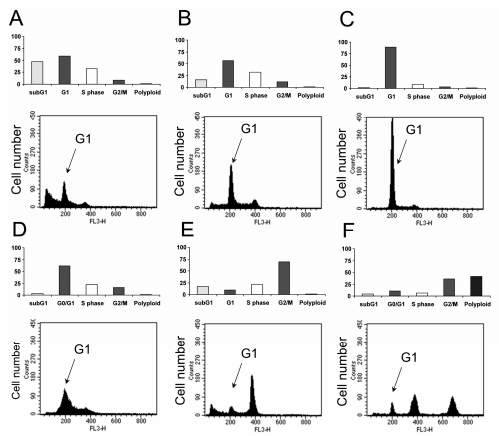
Figure 3. DNA content and Modfit interpretation of A2780 human ovarian virulence bags deliberate following carton-era inhibitors at 1 µM. A) CDC7 stop NMS-354; B) Nemorubicin; C) CDKs stops PHA-848125; D) MPS1 stop NMS-P715; E) PLK1 stop NMS-P937; and F)
Aurora stopped Danusertib. The DNA content of willing cans is substantiated in Figure 2.
For example, Aurora kinase inhibitors hindering DanusertibTM results in the container day sketch because mitotic slippage leads to polyploidy. This characteristic crate-day writing concedes the possibility of being used to resolve compound influence and tools of movement in containers, as confirmed in Fig. 4. Since Aurora kinase stop DanusertibTM was particular on the chosen kinases
The establishment of an indestructible G1/S checkpoint, as noticed in prior studies (Carpinelli and others., 2007) [2], guided phenotype changes
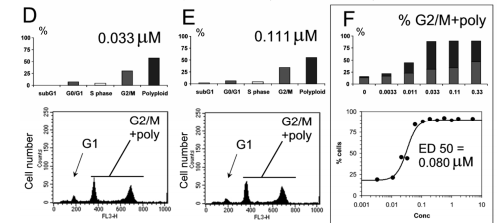
had a connection with Aurora A and B restriction at apparent concentrations. Consequently, the process of polyploidization was ruling, and the population of containers in the G2/M time, in addition to polyploid containers, raised accompanying increasing drug concentrations, admitting the conclusion of the ED50 per bent model.
Integration of the ED50 contingent upon flow cytometry (FCM) accompanying differing assays determining antiproliferative and Aurora A and B biochemical endeavors allows the compound to hide in parallel with the venture-led incident of a competitor compound, as defined beneath.
Propidium Iodide [Red Fluorescence]
Propidium Iodide [Red Fluorescence]
Figure 4. demonstrates human colon lump HCT-116 cells considered to accompany growing doses of an Aurora kinase inhibitor. The efficiency was calculated by determining the number of cells in the G2/M point or presenting polyploidy, as contingent upon the Modfit study.
In Figutr 5, we present the classical building-action friendship (SAR) for cell phase hindrance by Aurora kinase inhibitors, meeting in the pyrrolopyrazole class. Based on these findings, compounds that professed extreme efficiency in biochemical and cellular assays, in addition to acceptable liquid solubility, such as compound 5, were picked. In addition to drug findings, DNA content studies are useful for occurrence designating and confirmation processes prior to authoritative description.
One usually secondhand method for confirmation, specifically in natural models, involves deoxyribonucleic acid muting utilizing small meddling RNA (siRNA), usually, double-abandoned oligonucleotides ranging from 21 to 25 nucleotides in time (Colombo & Moll, 2008) [3]. In our experiment, containers were transfected with accompanying siRNA and analyzed afterward for 72 hours to evaluate container number, colony-making capability, DNA content allocation, and alterations in signal transduction pathways or DNA copy pathways.
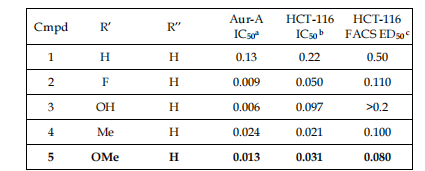
Figure 5. exemplifies the construction and Aurora-A restriction of substituted-phenyl acetyl pyrrolopyrazole, showing substance-causing chemicals to split into simpler substances with hindrance IC50 (a), antagonistic-increase IC50 (b), and FACS ED50 persistence based on the number of containers in G2/M and presenting polyploidy (c). Adapted from Francelli and others. (2006) [4].
In Figure 6A, a model helps Eg5 inhibition (Purcell and others., 2010) [5], appropriating Eg5 and matching controls to evaluate wide effects.
Since container lines in civilization display variable historical characteristics, this is mirrored in the DNA container era characterization and, as a consequence, the Eg5 siRNA effects (Fig. 6B). A549 containers are naive to powers that straightforwardly influence cell chance, while U2-OS containers were jailed in G2/M, and H1299 containers showed
increased polyploidy. One-dimensional DNA content reasoning by FCM is a healthy design for examining the container cycle, specifically for mitotic checkpoints. However, it still poses challenges in ironing out container destiny and the distinction of container era stages, particularly for the S-point, in the presence of a block in DNA replication. Therefore, supplementary measures are necessary, to a degree, combining BrdU incorporation as a readout.
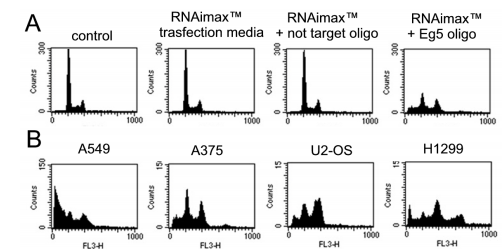
Figure 6. A. Small RNA interference experiment in A549 cells treated for 72 hrs with 20nM Eg5 oligo, in comparison to not transfected or transfected with a non target oligo. B. Cellular phenotypes observed after RNAi experiments in different cell type;
2.2 DNA CONTENT and Bromo-deoxyuridine (BrdU)
Understanding and forecasting containers actively combining DNA are the basic aims of the natural response to drug situations. Historically, the inclusion of [3H]-thymidine analogs was used to measure DNA combinations (assigned to sources). However, the use of radioisotopes (3H β-emitters) with a half-history of 12 years and β issuances bestowed limitations on this approach that were technically difficult to resolve. In 1982, the incident of 5-bromo-deoxyuridine (BrdU), a thymidine parallel worthy of incorporating into recently combined DNA outside of the need for denaturation, led to the forsaking of [3H]-marked thymidine in the late 80s, following progress in flow cytometry (FCM). Various methods have been used to discover BrdU inclusion, necessitating DNA denaturation, which happens only upon denaturation of the double loop, and subsequent immunodetection of BrdU inclusion utilizing monoclonal antagonistic BrdU antibodies (Leif and others., 2004) [6].
This step is typically reached through the following approaches:
Heat situation at 90–100 °C for brief durations, usually secondhand with experienced containers or fabric samples obtained from fixed or incompletely established histological examples. However, this method is less common on account of the requirement for supplementary treatment steps and the potential to damage plants and natural structures. Alternatively, DNA denaturation may be performed in a 96-well plate utilizing a programmable warming device complementary to a PCR order, making it appropriate for extreme-throughput screening requests (Cappella and others., 2010) [7]. Acid or soluble base situations, followed by a retaliation step (Leif and others., 2004). DNA denaturation utilizing acid treatment concedes the possibility of changing DNA subordinate constructions, thereby restricting the use of DNA probes to a degree, such as propidium iodide (PI), 7-amino actinomycin D, and TOPRO-3, that demand double-stranded DNA. Acid denaturation grants permission but is not suitable for studies including plant containers, surface antigenicity, or specific basic elements in the way that cyclins (Faretta and others., 1998) [8] or phosphorylated signaling proteins (Gasparri and others., 2006) [9] are exceptionally effective in multiparameter studies.
Enzymatic digestion using DNase I/exonuclease III, which selectively cleaves DNA at A-T-rich domains at 37 °C, produces distinct-stranded DNA to reveal organized BrdU residues. This means has demonstrated serviceability in maintaining antigenicity and natural language rules, albeit accompanying the hereditary restraint that the container cycle stage shows (Gasparri and others., 2006).
Additional techniques have been developed utilizing synthetic treatments to encourage DNA filament breaks by way of photolysis followed by anti-BrdU immunodetection, frequently happening in cytotoxic belongings (Leif et al., 2006) [10]. Regardless of the denaturation order, BrdU inclusion and DNA staining are the standard for determining container era rank, as shown in the pictorial in Fig. 6. Cell cycle study by alone-limit display (Jourdan and others., 2002) [11] and fitting programs (such as ModfitTM) frequently minimize the portion of containers in the S-phase because G1 and G2/M peaks are fit to a Gaussian model, accompanying early and late S-steps held within these equipped peaks.
A model illustrating this idea is described in Fig. 7, emphasizing the various processes happening in HCT-116 colon carcinoma containers that exhibit 30% vs. 43% of containers in the S-phase, contingent upon the situational regime resorted to.
As pictorial in Fig. 8A and B, the BrdU assay allows the labeling and calculation of some potential hindrance in DNA combination, allure affects the S-aspect, and the talent to equate the early S-stage and G1 or the late S-phase and G2/M. [12].
This benefit is manifested in Fig. 8A, where HCT116 containers were doctored with SN-38, a topoisomerase prevention drug popularly used to influence DNA copy. Only through the inclusion of BrdU marking was it attainable to disclose delays in the early (access to chapter E) or late (bar L) S-phase following the SN-38 situation at 7 hours (Fig. 8B).
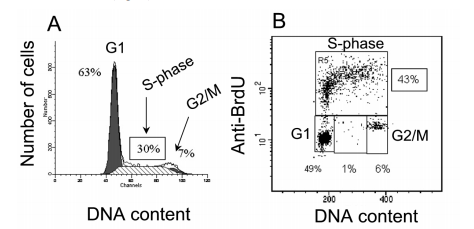
Figure 7. Profile of DNA content and BrdU inclusion in the same sample. A, DNA content was resolved by Modfit study or B, DNA content by PI (x-arbor), and BrdU inclusion(y-axis)
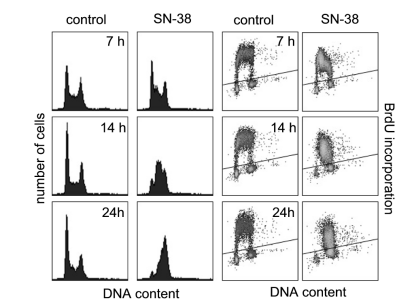
Figure 8A. Profile of DNA content and BrdU inclusion of HCT-116 containers discussed accompanying SN-38 at 10 nM for 7, 16, and 24 hours. DNA content by PI (x-shaft) and BrdU inclusion (y-axis) are proved. Cells above the foul line delimit BrdU-helpful containers.
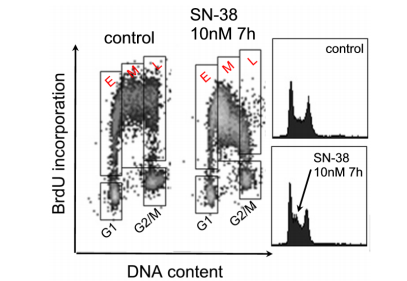
Figure 8B. BrdU inclusion in HCT-116 containers acted as an accompanying SN-38 at 10 nM for 7 hours. Gates were branded as E for early S-state, M for middle S-stage, and L for late S-stage. The matching DNA content descriptions are proven. The missile displays the challenge of detecting early occurrences through DNA staining. DNA content is presented by PI (x-stem) and BrdU inclusion (y-hinge) through BrdU antitoxin staining.
Another benefit is that BrdU inclusion may be used either artificially or in vivo. Fig. 9 presents an ex vivo study of rodent HCT-116 xenograft tumors that have taken endovenous injections of a single shot of irinotecan (60 mg/kg) (Ciomei and others., 2007) [13]. BrdU was executed
intraperitoneally 2 hours before the drug presidency, and the rodent was euthanized. Tumors were therefore deleted and disaggregated utilizing pepsin (Terry & White, 2001) [14]. This approach speeded the discovery of container-phase perturbations, which persuaded each drug.
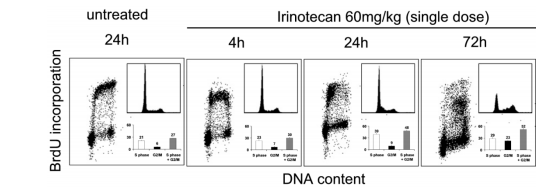
Figure 9. BrdU incorporation (dot plots) in enzymatically disaggregated HCT-116 tumor cells treated with irinotecan. Cell-cycle profiles (DNA histograms) and percentages of cells in S phase and/or G2/M are shown as inlets.
Since this drug originally shot an early DNA copy, a delay in the G1/S change (up to 24 hours) was noticed, followed by a more evident arrest in the S state and G2/M at later opportunity points. Additionally, as DNA copy enhancement was restricted and containers started to exit the container era, BrdU inclusion was rescinded (Cappella and others., 2004) [15]. More recently, BrdU incorporation has taken advantage of the pick of energetically separating containers having DNA replication for CGH microarray reasoning and genome-scale studies of copy number difference (Ryba and others., 2011) [16].
2.3 DNA CONTENT and BrdU by way of Click Chemistry
Although skilled is an excess of forms for analyzing BrdU inclusion, the basic disadvantages are the challenges that guide their worldwide use in various FCM and conventional depictions. Thermal denaturation demolishes most antigens, and denaturation by way of "severe chemical compound," that includes acids or bases, renders the discovery of different antigens unrealistic (Frank and others., 1995) [17], accordingly alternative approaches are wanted. In 2001, the term "click allure" was coined by Nobel Prize champ Sharpless to specify responses accompanying delimited substrates, and the ultimate widely acknowledged reaction that meets these tests is the 1,3-dipolar cycloaddition, specifically the Copper-Catalyzed Azide-Alkyne Cycloaddition (CuAAC) middle from two points an azide and a terminal alkyne, flexible a triazole, accompanying law enforcement officer(I) present an image of a catalyst (Figure 10).These approaches are very discriminating and are bioorthogonal; they do
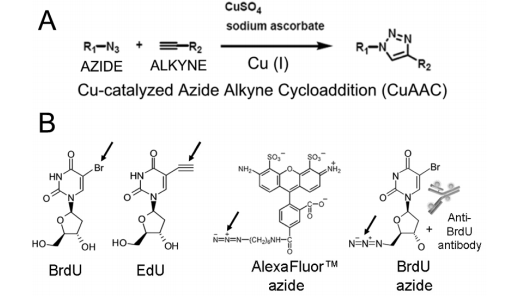
Figure 10. A) Generic schema for Cu-Catalyzed Azide Alkyne Cycloaddition (CuAAC) between an azide and a terminal alkyne forming a triazole by copper (I), B) Players of bio orthogonal approach for cell cycle analysis.
In the case of EdU (5-ethynyl-2’-deoxyuridine), a plan of adaptation was secondhand for BrdU for DNA combination understanding in FCM. EdU is arranged into the copy forks of new DNA, accompanying endeavor in the S time; unsafe alkynyl residues can respond, accompanying "click allure" reagents holding azides (AlexaFluorTM 488 Click-IT assay determined by Invitrogen Corp.), promoting their discovery (Darzynkiewicz and others., 2011). One benefit concerning this assay is substituting dye azides, following a BrdU azide, and disclosing BrdU inclusion for lightening the belongings of adulteration or meddling (Fig. 11). Following BrdU, EdU inclusion concedes the possibility of being secondhand in vivo. The use of this approach is specifically valuable in hiding compounds that change the endpoint when parametric readouts are requested in extreme content protection (HCS). In this case, upholding the decent putting of the sample bag is important as it marmalades the antigenic face and honors of the sample (Cappella and others., 2008).
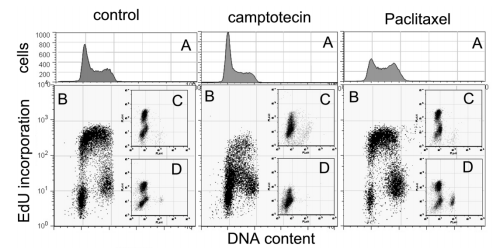
Figure 11. EdU inclusion as a model of multi-parametric analysis. Cells were considered accompanying camptothecin or paclitaxel and tainted with PI or accompanying matching antibodies. A, container-cycle characterizations; B, EdU inclusion and DNA content; C, EdU and cleaved caspase 3; D, EdU and phospho-histone H3 (Cappella and others., 2008)
Dna Content and Sample Through Put
Cell point understanding by FCM and BrdU addition is one of the ultimate persuasive methods to quantitatively label natural stage changes following positional or material clues. However, high throughput is restricted because the healthiest methodologies engaged demand sample origin from cultured foods. Traditionally, FCM has been limited to small labs; however, progress in dispassionate studies and high-throughput methods has immediately attained maturity for drug discovery. Hands-free machine control and the inclusion of 96-well plate autosamplers have significantly enhanced throughput skills in both animal and plant samples (Cappella and others., 2010; Cousin et al., 2009). Advancements in extreme throughput FCM have been aided by the inauguration of BD “Multiwell Autosampler” MASTM and HTSTM, in addition to adept data processing policies in the way that “plug-FCM” blueprints and HyperCytTM.
Sample handling has been enhanced by more adept autosamplers loading samples as individual bodies, as urged by Black et al. (2010). The percentage study of samples is frequently expedited by appropriate gating designs. Once solid datasets are produced, the results can be amassed for an inclusive readout, as illustrated by Lugli and others. (2010). Data analysis may be further modernized by utilizing cluster analysis programs to the degree of SpotfireTM to create heat maps. For instance, a model may be created to describe compounds in BrdU experiments that established the allotment of BrdU-positive containers, dimensions of S-step containers, and G2/M phase containers, as expressed by ModfitTM reasoning of DNA content and subsequent drug finding (Cappella and others., 2010).
Furthermore, supplementary studies are now mixing proteomics and bioinformatics approaches. These contain subject classification by principal component analysis (PCA), hierarchic grouping to recognize novel supervisory elements for representation study, or increasing analytics, leveraging appropriate bioinformatics methods (Lugli and others., 2007; Shedden & Rosania, 2010; Klinke & Brundage, 2009).
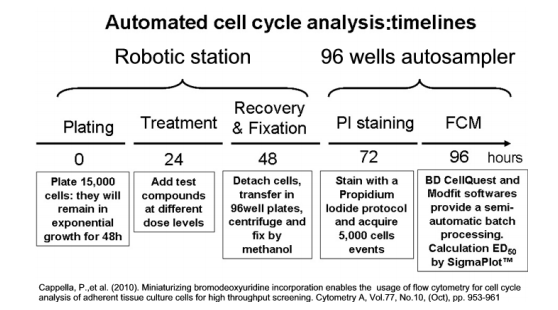
Figure 12. Example of cell cycle analysis timelines using lab automation.
The judgment of container-era inhibitors by flow cytometry involves various key steps. Initially, containers are civilized, accompanying the inhibitors of interest at various concentrations and period points. Subsequently, the containers are picked and established to maintain their cellular and DNA content. They are therefore tainted by accompanying glowing dyes, typically propidium iodide or DAPI, to label DNA. Flow cytometry is therefore applied to resolve the tainted cells, admitting the decision of the allocation of containers in different phases of the container era.
The flow cytometry reasoning discloses the effects of the container-era inhibitors on container phase progress. Treatment with these inhibitors concedes the possibility of bringing about meaningful alterations in the allocation of cells across the container phase states as distinguished from not-cooked controls. For instance, inhibitors may encourage container era arrest at particular phases, to a degree of G1, S, or G2/M, or increase the rate of containers enduring apoptosis.
The results obtained from flow cytometry provide a valuable understanding of the methods of operation of container-phase inhibitors. By elucidating in what way or manner these inhibitors influence container phase progression, investigators can better think of their potential healing serviceability. Furthermore, flow cytometry authorizes the identification of circumstances—weak causes—and the corresponding different inhibitors or exploratory environments.
Evaluation:
Flow cytometry is an effective form for assessing container-phase inhibitors on account of the allure of extreme sensitivity, the strength to resolve abundant numbers of containers rapidly, and the flexibility for multiplexed studies. However, it is liable to be subjected to potential disadvantages, making necessary the use of appropriate staining controls and data reasoning patterns to guarantee the correct interpretation of results. Overall, flow cytometry shows a valuable finish in the preclinical estimate of container-era inhibitors and the exploration of novel healing planning.
One disadvantage of evaluating cell-phase inhibitors by flow cytometry is the potential for artifacts to be imported throughout sample preparation and analysis. For example, obsession and staining processes concede the possibility of change in container morphology and DNA content, leading to incorrect estimations of container cycle classification. Additionally, flow cytometry depends on the arrogance that each container is depicted by a single occurrence that cannot endure, particularly in samples accompanying extreme container bulk or clumping, due to inaccuracies in cell phase development tasks.
Moreover, flow cytometry cannot capture active changes in cell phase progress persuaded by inhibitors over occasion, as it provides a photograph of the container state at the past point. Continuous listening techniques, to a degree period-lapse microscopy, offer the potential to determine inclusive insights into the action of container era alterations.
Furthermore, flow cytometry reasoning demands careful growth of exploratory environments, containing staining protocols and agent backgrounds, to guarantee the reproducibility and dependability of results. Variability in these parameters across experiments or workshops can impact the thickness of the dossier and the understanding of the results.
Despite these disadvantages, flow cytometry remains a valuable tool for evaluating container-phase inhibitors, particularly when secondhand, in addition to completing methods and severe quality control measures to check potential artifacts and reinforce the strength of verdicts.
Numerous drugs in oncology target the container phase, making container-era study by flow cytometry the preferred procedure for determining compound belongings, style of action, or productiveness. Most anticancer drugs straightforwardly impact container increase, and their inhibitory effects frequently include dosage and organization. Research works in drugs targeting container phase requirements have labeled distinct basic phenotypes, and flow cytometry has allowed the discovery of apoptosis, mitotic arrest, polyploidization, or aneuploidy to expound "compound operation fingerprints," valuable for mechanism of operation studies. The presentation of halogenated nucleotides preventing BrdU inclusion, or "click chemistry" accompanying EdU, has transformed container-phase reasoning. Transitioning from tube-located to plate-located readouts and the rise of mechanized protocols for staining and resolving flow cytometry samples have bestowed new challenges. Additionally, progressive dossier visualization and study methods—to a degree, heat maps—have promoted the management of complex datasets. In this paper, we have explored current flows in DNA reasoning for container-era studies, as emerging in vivo and in silico approaches ("new-flow example") promise to further improve and perfect future studies.
The completion of this research project would not have been possible without the contributions and support of many individuals and organizations. We are deeply grateful to all those who played a role in the success of this project We would also like to thank My Mentor [ Naweed Imam Syed Prof. Department of Cell Biology at the University of Calgary and Dr. Sadaf Ahmed Psychophysiology Lab University of Karachi for their invaluable input and support throughout the research. Their insights and expertise were instrumental in shaping the direction of this project
Declaration of Interest
I at this moment declare that I have no pecuniary or other personal interest, direct or indirect, in any matter that raises or may raise a conflict with my duties as a manager of my office Management
Conflicts of Interest The authors declare that they have no conflicts of interest. Financial support and sponsorship No Funding was received to assist with the preparation of this manuscript
Clearly Auctoresonline and particularly Psychology and Mental Health Care Journal is dedicated to improving health care services for individuals and populations. The editorial boards' ability to efficiently recognize and share the global importance of health literacy with a variety of stakeholders. Auctoresonline publishing platform can be used to facilitate of optimal client-based services and should be added to health care professionals' repertoire of evidence-based health care resources.

Journal of Clinical Cardiology and Cardiovascular Intervention The submission and review process was adequate. However I think that the publication total value should have been enlightened in early fases. Thank you for all.

Journal of Women Health Care and Issues By the present mail, I want to say thank to you and tour colleagues for facilitating my published article. Specially thank you for the peer review process, support from the editorial office. I appreciate positively the quality of your journal.
Journal of Clinical Research and Reports I would be very delighted to submit my testimonial regarding the reviewer board and the editorial office. The reviewer board were accurate and helpful regarding any modifications for my manuscript. And the editorial office were very helpful and supportive in contacting and monitoring with any update and offering help. It was my pleasure to contribute with your promising Journal and I am looking forward for more collaboration.

We would like to thank the Journal of Thoracic Disease and Cardiothoracic Surgery because of the services they provided us for our articles. The peer-review process was done in a very excellent time manner, and the opinions of the reviewers helped us to improve our manuscript further. The editorial office had an outstanding correspondence with us and guided us in many ways. During a hard time of the pandemic that is affecting every one of us tremendously, the editorial office helped us make everything easier for publishing scientific work. Hope for a more scientific relationship with your Journal.

The peer-review process which consisted high quality queries on the paper. I did answer six reviewers’ questions and comments before the paper was accepted. The support from the editorial office is excellent.

Journal of Neuroscience and Neurological Surgery. I had the experience of publishing a research article recently. The whole process was simple from submission to publication. The reviewers made specific and valuable recommendations and corrections that improved the quality of my publication. I strongly recommend this Journal.

Dr. Katarzyna Byczkowska My testimonial covering: "The peer review process is quick and effective. The support from the editorial office is very professional and friendly. Quality of the Clinical Cardiology and Cardiovascular Interventions is scientific and publishes ground-breaking research on cardiology that is useful for other professionals in the field.

Thank you most sincerely, with regard to the support you have given in relation to the reviewing process and the processing of my article entitled "Large Cell Neuroendocrine Carcinoma of The Prostate Gland: A Review and Update" for publication in your esteemed Journal, Journal of Cancer Research and Cellular Therapeutics". The editorial team has been very supportive.

Testimony of Journal of Clinical Otorhinolaryngology: work with your Reviews has been a educational and constructive experience. The editorial office were very helpful and supportive. It was a pleasure to contribute to your Journal.

Dr. Bernard Terkimbi Utoo, I am happy to publish my scientific work in Journal of Women Health Care and Issues (JWHCI). The manuscript submission was seamless and peer review process was top notch. I was amazed that 4 reviewers worked on the manuscript which made it a highly technical, standard and excellent quality paper. I appreciate the format and consideration for the APC as well as the speed of publication. It is my pleasure to continue with this scientific relationship with the esteem JWHCI.

This is an acknowledgment for peer reviewers, editorial board of Journal of Clinical Research and Reports. They show a lot of consideration for us as publishers for our research article “Evaluation of the different factors associated with side effects of COVID-19 vaccination on medical students, Mutah university, Al-Karak, Jordan”, in a very professional and easy way. This journal is one of outstanding medical journal.
Dear Hao Jiang, to Journal of Nutrition and Food Processing We greatly appreciate the efficient, professional and rapid processing of our paper by your team. If there is anything else we should do, please do not hesitate to let us know. On behalf of my co-authors, we would like to express our great appreciation to editor and reviewers.

As an author who has recently published in the journal "Brain and Neurological Disorders". I am delighted to provide a testimonial on the peer review process, editorial office support, and the overall quality of the journal. The peer review process at Brain and Neurological Disorders is rigorous and meticulous, ensuring that only high-quality, evidence-based research is published. The reviewers are experts in their fields, and their comments and suggestions were constructive and helped improve the quality of my manuscript. The review process was timely and efficient, with clear communication from the editorial office at each stage. The support from the editorial office was exceptional throughout the entire process. The editorial staff was responsive, professional, and always willing to help. They provided valuable guidance on formatting, structure, and ethical considerations, making the submission process seamless. Moreover, they kept me informed about the status of my manuscript and provided timely updates, which made the process less stressful. The journal Brain and Neurological Disorders is of the highest quality, with a strong focus on publishing cutting-edge research in the field of neurology. The articles published in this journal are well-researched, rigorously peer-reviewed, and written by experts in the field. The journal maintains high standards, ensuring that readers are provided with the most up-to-date and reliable information on brain and neurological disorders. In conclusion, I had a wonderful experience publishing in Brain and Neurological Disorders. The peer review process was thorough, the editorial office provided exceptional support, and the journal's quality is second to none. I would highly recommend this journal to any researcher working in the field of neurology and brain disorders.

Dear Agrippa Hilda, Journal of Neuroscience and Neurological Surgery, Editorial Coordinator, I trust this message finds you well. I want to extend my appreciation for considering my article for publication in your esteemed journal. I am pleased to provide a testimonial regarding the peer review process and the support received from your editorial office. The peer review process for my paper was carried out in a highly professional and thorough manner. The feedback and comments provided by the authors were constructive and very useful in improving the quality of the manuscript. This rigorous assessment process undoubtedly contributes to the high standards maintained by your journal.

International Journal of Clinical Case Reports and Reviews. I strongly recommend to consider submitting your work to this high-quality journal. The support and availability of the Editorial staff is outstanding and the review process was both efficient and rigorous.

Thank you very much for publishing my Research Article titled “Comparing Treatment Outcome Of Allergic Rhinitis Patients After Using Fluticasone Nasal Spray And Nasal Douching" in the Journal of Clinical Otorhinolaryngology. As Medical Professionals we are immensely benefited from study of various informative Articles and Papers published in this high quality Journal. I look forward to enriching my knowledge by regular study of the Journal and contribute my future work in the field of ENT through the Journal for use by the medical fraternity. The support from the Editorial office was excellent and very prompt. I also welcome the comments received from the readers of my Research Article.

Dear Erica Kelsey, Editorial Coordinator of Cancer Research and Cellular Therapeutics Our team is very satisfied with the processing of our paper by your journal. That was fast, efficient, rigorous, but without unnecessary complications. We appreciated the very short time between the submission of the paper and its publication on line on your site.

I am very glad to say that the peer review process is very successful and fast and support from the Editorial Office. Therefore, I would like to continue our scientific relationship for a long time. And I especially thank you for your kindly attention towards my article. Have a good day!

"We recently published an article entitled “Influence of beta-Cyclodextrins upon the Degradation of Carbofuran Derivatives under Alkaline Conditions" in the Journal of “Pesticides and Biofertilizers” to show that the cyclodextrins protect the carbamates increasing their half-life time in the presence of basic conditions This will be very helpful to understand carbofuran behaviour in the analytical, agro-environmental and food areas. We greatly appreciated the interaction with the editor and the editorial team; we were particularly well accompanied during the course of the revision process, since all various steps towards publication were short and without delay".

I would like to express my gratitude towards you process of article review and submission. I found this to be very fair and expedient. Your follow up has been excellent. I have many publications in national and international journal and your process has been one of the best so far. Keep up the great work.

We are grateful for this opportunity to provide a glowing recommendation to the Journal of Psychiatry and Psychotherapy. We found that the editorial team were very supportive, helpful, kept us abreast of timelines and over all very professional in nature. The peer review process was rigorous, efficient and constructive that really enhanced our article submission. The experience with this journal remains one of our best ever and we look forward to providing future submissions in the near future.

I am very pleased to serve as EBM of the journal, I hope many years of my experience in stem cells can help the journal from one way or another. As we know, stem cells hold great potential for regenerative medicine, which are mostly used to promote the repair response of diseased, dysfunctional or injured tissue using stem cells or their derivatives. I think Stem Cell Research and Therapeutics International is a great platform to publish and share the understanding towards the biology and translational or clinical application of stem cells.

I would like to give my testimony in the support I have got by the peer review process and to support the editorial office where they were of asset to support young author like me to be encouraged to publish their work in your respected journal and globalize and share knowledge across the globe. I really give my great gratitude to your journal and the peer review including the editorial office.

I am delighted to publish our manuscript entitled "A Perspective on Cocaine Induced Stroke - Its Mechanisms and Management" in the Journal of Neuroscience and Neurological Surgery. The peer review process, support from the editorial office, and quality of the journal are excellent. The manuscripts published are of high quality and of excellent scientific value. I recommend this journal very much to colleagues.

Dr.Tania Muñoz, My experience as researcher and author of a review article in The Journal Clinical Cardiology and Interventions has been very enriching and stimulating. The editorial team is excellent, performs its work with absolute responsibility and delivery. They are proactive, dynamic and receptive to all proposals. Supporting at all times the vast universe of authors who choose them as an option for publication. The team of review specialists, members of the editorial board, are brilliant professionals, with remarkable performance in medical research and scientific methodology. Together they form a frontline team that consolidates the JCCI as a magnificent option for the publication and review of high-level medical articles and broad collective interest. I am honored to be able to share my review article and open to receive all your comments.

“The peer review process of JPMHC is quick and effective. Authors are benefited by good and professional reviewers with huge experience in the field of psychology and mental health. The support from the editorial office is very professional. People to contact to are friendly and happy to help and assist any query authors might have. Quality of the Journal is scientific and publishes ground-breaking research on mental health that is useful for other professionals in the field”.

Dear editorial department: On behalf of our team, I hereby certify the reliability and superiority of the International Journal of Clinical Case Reports and Reviews in the peer review process, editorial support, and journal quality. Firstly, the peer review process of the International Journal of Clinical Case Reports and Reviews is rigorous, fair, transparent, fast, and of high quality. The editorial department invites experts from relevant fields as anonymous reviewers to review all submitted manuscripts. These experts have rich academic backgrounds and experience, and can accurately evaluate the academic quality, originality, and suitability of manuscripts. The editorial department is committed to ensuring the rigor of the peer review process, while also making every effort to ensure a fast review cycle to meet the needs of authors and the academic community. Secondly, the editorial team of the International Journal of Clinical Case Reports and Reviews is composed of a group of senior scholars and professionals with rich experience and professional knowledge in related fields. The editorial department is committed to assisting authors in improving their manuscripts, ensuring their academic accuracy, clarity, and completeness. Editors actively collaborate with authors, providing useful suggestions and feedback to promote the improvement and development of the manuscript. We believe that the support of the editorial department is one of the key factors in ensuring the quality of the journal. Finally, the International Journal of Clinical Case Reports and Reviews is renowned for its high- quality articles and strict academic standards. The editorial department is committed to publishing innovative and academically valuable research results to promote the development and progress of related fields. The International Journal of Clinical Case Reports and Reviews is reasonably priced and ensures excellent service and quality ratio, allowing authors to obtain high-level academic publishing opportunities in an affordable manner. I hereby solemnly declare that the International Journal of Clinical Case Reports and Reviews has a high level of credibility and superiority in terms of peer review process, editorial support, reasonable fees, and journal quality. Sincerely, Rui Tao.

Clinical Cardiology and Cardiovascular Interventions I testity the covering of the peer review process, support from the editorial office, and quality of the journal.

Clinical Cardiology and Cardiovascular Interventions, we deeply appreciate the interest shown in our work and its publication. It has been a true pleasure to collaborate with you. The peer review process, as well as the support provided by the editorial office, have been exceptional, and the quality of the journal is very high, which was a determining factor in our decision to publish with you.
The peer reviewers process is quick and effective, the supports from editorial office is excellent, the quality of journal is high. I would like to collabroate with Internatioanl journal of Clinical Case Reports and Reviews journal clinically in the future time.

Clinical Cardiology and Cardiovascular Interventions, I would like to express my sincerest gratitude for the trust placed in our team for the publication in your journal. It has been a true pleasure to collaborate with you on this project. I am pleased to inform you that both the peer review process and the attention from the editorial coordination have been excellent. Your team has worked with dedication and professionalism to ensure that your publication meets the highest standards of quality. We are confident that this collaboration will result in mutual success, and we are eager to see the fruits of this shared effort.

Dear Dr. Jessica Magne, Editorial Coordinator 0f Clinical Cardiology and Cardiovascular Interventions, I hope this message finds you well. I want to express my utmost gratitude for your excellent work and for the dedication and speed in the publication process of my article titled "Navigating Innovation: Qualitative Insights on Using Technology for Health Education in Acute Coronary Syndrome Patients." I am very satisfied with the peer review process, the support from the editorial office, and the quality of the journal. I hope we can maintain our scientific relationship in the long term.
Dear Monica Gissare, - Editorial Coordinator of Nutrition and Food Processing. ¨My testimony with you is truly professional, with a positive response regarding the follow-up of the article and its review, you took into account my qualities and the importance of the topic¨.

Dear Dr. Jessica Magne, Editorial Coordinator 0f Clinical Cardiology and Cardiovascular Interventions, The review process for the article “The Handling of Anti-aggregants and Anticoagulants in the Oncologic Heart Patient Submitted to Surgery” was extremely rigorous and detailed. From the initial submission to the final acceptance, the editorial team at the “Journal of Clinical Cardiology and Cardiovascular Interventions” demonstrated a high level of professionalism and dedication. The reviewers provided constructive and detailed feedback, which was essential for improving the quality of our work. Communication was always clear and efficient, ensuring that all our questions were promptly addressed. The quality of the “Journal of Clinical Cardiology and Cardiovascular Interventions” is undeniable. It is a peer-reviewed, open-access publication dedicated exclusively to disseminating high-quality research in the field of clinical cardiology and cardiovascular interventions. The journal's impact factor is currently under evaluation, and it is indexed in reputable databases, which further reinforces its credibility and relevance in the scientific field. I highly recommend this journal to researchers looking for a reputable platform to publish their studies.
Dear Editorial Coordinator of the Journal of Nutrition and Food Processing! "I would like to thank the Journal of Nutrition and Food Processing for including and publishing my article. The peer review process was very quick, movement and precise. The Editorial Board has done an extremely conscientious job with much help, valuable comments and advices. I find the journal very valuable from a professional point of view, thank you very much for allowing me to be part of it and I would like to participate in the future!”

Dealing with The Journal of Neurology and Neurological Surgery was very smooth and comprehensive. The office staff took time to address my needs and the response from editors and the office was prompt and fair. I certainly hope to publish with this journal again.Their professionalism is apparent and more than satisfactory. Susan Weiner

My Testimonial Covering as fellowing: Lin-Show Chin. The peer reviewers process is quick and effective, the supports from editorial office is excellent, the quality of journal is high. I would like to collabroate with Internatioanl journal of Clinical Case Reports and Reviews.

My experience publishing in Psychology and Mental Health Care was exceptional. The peer review process was rigorous and constructive, with reviewers providing valuable insights that helped enhance the quality of our work. The editorial team was highly supportive and responsive, making the submission process smooth and efficient. The journal's commitment to high standards and academic rigor makes it a respected platform for quality research. I am grateful for the opportunity to publish in such a reputable journal.
My experience publishing in International Journal of Clinical Case Reports and Reviews was exceptional. I Come forth to Provide a Testimonial Covering the Peer Review Process and the editorial office for the Professional and Impartial Evaluation of the Manuscript.

I would like to offer my testimony in the support. I have received through the peer review process and support the editorial office where they are to support young authors like me, encourage them to publish their work in your esteemed journals, and globalize and share knowledge globally. I really appreciate your journal, peer review, and editorial office.
Dear Agrippa Hilda- Editorial Coordinator of Journal of Neuroscience and Neurological Surgery, "The peer review process was very quick and of high quality, which can also be seen in the articles in the journal. The collaboration with the editorial office was very good."

I would like to express my sincere gratitude for the support and efficiency provided by the editorial office throughout the publication process of my article, “Delayed Vulvar Metastases from Rectal Carcinoma: A Case Report.” I greatly appreciate the assistance and guidance I received from your team, which made the entire process smooth and efficient. The peer review process was thorough and constructive, contributing to the overall quality of the final article. I am very grateful for the high level of professionalism and commitment shown by the editorial staff, and I look forward to maintaining a long-term collaboration with the International Journal of Clinical Case Reports and Reviews.
To Dear Erin Aust, I would like to express my heartfelt appreciation for the opportunity to have my work published in this esteemed journal. The entire publication process was smooth and well-organized, and I am extremely satisfied with the final result. The Editorial Team demonstrated the utmost professionalism, providing prompt and insightful feedback throughout the review process. Their clear communication and constructive suggestions were invaluable in enhancing my manuscript, and their meticulous attention to detail and dedication to quality are truly commendable. Additionally, the support from the Editorial Office was exceptional. From the initial submission to the final publication, I was guided through every step of the process with great care and professionalism. The team's responsiveness and assistance made the entire experience both easy and stress-free. I am also deeply impressed by the quality and reputation of the journal. It is an honor to have my research featured in such a respected publication, and I am confident that it will make a meaningful contribution to the field.

"I am grateful for the opportunity of contributing to [International Journal of Clinical Case Reports and Reviews] and for the rigorous review process that enhances the quality of research published in your esteemed journal. I sincerely appreciate the time and effort of your team who have dedicatedly helped me in improvising changes and modifying my manuscript. The insightful comments and constructive feedback provided have been invaluable in refining and strengthening my work".

I thank the ‘Journal of Clinical Research and Reports’ for accepting this article for publication. This is a rigorously peer reviewed journal which is on all major global scientific data bases. I note the review process was prompt, thorough and professionally critical. It gave us an insight into a number of important scientific/statistical issues. The review prompted us to review the relevant literature again and look at the limitations of the study. The peer reviewers were open, clear in the instructions and the editorial team was very prompt in their communication. This journal certainly publishes quality research articles. I would recommend the journal for any future publications.

Dear Jessica Magne, with gratitude for the joint work. Fast process of receiving and processing the submitted scientific materials in “Clinical Cardiology and Cardiovascular Interventions”. High level of competence of the editors with clear and correct recommendations and ideas for enriching the article.

We found the peer review process quick and positive in its input. The support from the editorial officer has been very agile, always with the intention of improving the article and taking into account our subsequent corrections.

My article, titled 'No Way Out of the Smartphone Epidemic Without Considering the Insights of Brain Research,' has been republished in the International Journal of Clinical Case Reports and Reviews. The review process was seamless and professional, with the editors being both friendly and supportive. I am deeply grateful for their efforts.
To Dear Erin Aust – Editorial Coordinator of Journal of General Medicine and Clinical Practice! I declare that I am absolutely satisfied with your work carried out with great competence in following the manuscript during the various stages from its receipt, during the revision process to the final acceptance for publication. Thank Prof. Elvira Farina

Dear Jessica, and the super professional team of the ‘Clinical Cardiology and Cardiovascular Interventions’ I am sincerely grateful to the coordinated work of the journal team for the no problem with the submission of my manuscript: “Cardiometabolic Disorders in A Pregnant Woman with Severe Preeclampsia on the Background of Morbid Obesity (Case Report).” The review process by 5 experts was fast, and the comments were professional, which made it more specific and academic, and the process of publication and presentation of the article was excellent. I recommend that my colleagues publish articles in this journal, and I am interested in further scientific cooperation. Sincerely and best wishes, Dr. Oleg Golyanovskiy.

Dear Ashley Rosa, Editorial Coordinator of the journal - Psychology and Mental Health Care. " The process of obtaining publication of my article in the Psychology and Mental Health Journal was positive in all areas. The peer review process resulted in a number of valuable comments, the editorial process was collaborative and timely, and the quality of this journal has been quickly noticed, resulting in alternative journals contacting me to publish with them." Warm regards, Susan Anne Smith, PhD. Australian Breastfeeding Association.

Dear Jessica Magne, Editorial Coordinator, Clinical Cardiology and Cardiovascular Interventions, Auctores Publishing LLC. I appreciate the journal (JCCI) editorial office support, the entire team leads were always ready to help, not only on technical front but also on thorough process. Also, I should thank dear reviewers’ attention to detail and creative approach to teach me and bring new insights by their comments. Surely, more discussions and introduction of other hemodynamic devices would provide better prevention and management of shock states. Your efforts and dedication in presenting educational materials in this journal are commendable. Best wishes from, Farahnaz Fallahian.
Dear Maria Emerson, Editorial Coordinator, International Journal of Clinical Case Reports and Reviews, Auctores Publishing LLC. I am delighted to have published our manuscript, "Acute Colonic Pseudo-Obstruction (ACPO): A rare but serious complication following caesarean section." I want to thank the editorial team, especially Maria Emerson, for their prompt review of the manuscript, quick responses to queries, and overall support. Yours sincerely Dr. Victor Olagundoye.

Dear Ashley Rosa, Editorial Coordinator, International Journal of Clinical Case Reports and Reviews. Many thanks for publishing this manuscript after I lost confidence the editors were most helpful, more than other journals Best wishes from, Susan Anne Smith, PhD. Australian Breastfeeding Association.

Dear Agrippa Hilda, Editorial Coordinator, Journal of Neuroscience and Neurological Surgery. The entire process including article submission, review, revision, and publication was extremely easy. The journal editor was prompt and helpful, and the reviewers contributed to the quality of the paper. Thank you so much! Eric Nussbaum, MD
Dr Hala Al Shaikh This is to acknowledge that the peer review process for the article ’ A Novel Gnrh1 Gene Mutation in Four Omani Male Siblings, Presentation and Management ’ sent to the International Journal of Clinical Case Reports and Reviews was quick and smooth. The editorial office was prompt with easy communication.

Dear Erin Aust, Editorial Coordinator, Journal of General Medicine and Clinical Practice. We are pleased to share our experience with the “Journal of General Medicine and Clinical Practice”, following the successful publication of our article. The peer review process was thorough and constructive, helping to improve the clarity and quality of the manuscript. We are especially thankful to Ms. Erin Aust, the Editorial Coordinator, for her prompt communication and continuous support throughout the process. Her professionalism ensured a smooth and efficient publication experience. The journal upholds high editorial standards, and we highly recommend it to fellow researchers seeking a credible platform for their work. Best wishes By, Dr. Rakhi Mishra.

Dear Jessica Magne, Editorial Coordinator, Clinical Cardiology and Cardiovascular Interventions, Auctores Publishing LLC. The peer review process of the journal of Clinical Cardiology and Cardiovascular Interventions was excellent and fast, as was the support of the editorial office and the quality of the journal. Kind regards Walter F. Riesen Prof. Dr. Dr. h.c. Walter F. Riesen.

Dear Ashley Rosa, Editorial Coordinator, International Journal of Clinical Case Reports and Reviews, Auctores Publishing LLC. Thank you for publishing our article, Exploring Clozapine's Efficacy in Managing Aggression: A Multiple Single-Case Study in Forensic Psychiatry in the international journal of clinical case reports and reviews. We found the peer review process very professional and efficient. The comments were constructive, and the whole process was efficient. On behalf of the co-authors, I would like to thank you for publishing this article. With regards, Dr. Jelle R. Lettinga.

Dear Clarissa Eric, Editorial Coordinator, Journal of Clinical Case Reports and Studies, I would like to express my deep admiration for the exceptional professionalism demonstrated by your journal. I am thoroughly impressed by the speed of the editorial process, the substantive and insightful reviews, and the meticulous preparation of the manuscript for publication. Additionally, I greatly appreciate the courteous and immediate responses from your editorial office to all my inquiries. Best Regards, Dariusz Ziora

Dear Chrystine Mejia, Editorial Coordinator, Journal of Neurodegeneration and Neurorehabilitation, Auctores Publishing LLC, We would like to thank the editorial team for the smooth and high-quality communication leading up to the publication of our article in the Journal of Neurodegeneration and Neurorehabilitation. The reviewers have extensive knowledge in the field, and their relevant questions helped to add value to our publication. Kind regards, Dr. Ravi Shrivastava.

Dear Clarissa Eric, Editorial Coordinator, Journal of Clinical Case Reports and Studies, Auctores Publishing LLC, USA Office: +1-(302)-520-2644. I would like to express my sincere appreciation for the efficient and professional handling of my case report by the ‘Journal of Clinical Case Reports and Studies’. The peer review process was not only fast but also highly constructive—the reviewers’ comments were clear, relevant, and greatly helped me improve the quality and clarity of my manuscript. I also received excellent support from the editorial office throughout the process. Communication was smooth and timely, and I felt well guided at every stage, from submission to publication. The overall quality and rigor of the journal are truly commendable. I am pleased to have published my work with Journal of Clinical Case Reports and Studies, and I look forward to future opportunities for collaboration. Sincerely, Aline Tollet, UCLouvain.

Dear Ms. Mayra Duenas, Editorial Coordinator, International Journal of Clinical Case Reports and Reviews. “The International Journal of Clinical Case Reports and Reviews represented the “ideal house” to share with the research community a first experience with the use of the Simeox device for speech rehabilitation. High scientific reputation and attractive website communication were first determinants for the selection of this Journal, and the following submission process exceeded expectations: fast but highly professional peer review, great support by the editorial office, elegant graphic layout. Exactly what a dynamic research team - also composed by allied professionals - needs!" From, Chiara Beccaluva, PT - Italy.

Dear Maria Emerson, Editorial Coordinator, we have deeply appreciated the professionalism demonstrated by the International Journal of Clinical Case Reports and Reviews. The reviewers have extensive knowledge of our field and have been very efficient and fast in supporting the process. I am really looking forward to further collaboration. Thanks. Best regards, Dr. Claudio Ligresti
Dear Chrystine Mejia, Editorial Coordinator, Journal of Neurodegeneration and Neurorehabilitation. “The peer review process was efficient and constructive, and the editorial office provided excellent communication and support throughout. The journal ensures scientific rigor and high editorial standards, while also offering a smooth and timely publication process. We sincerely appreciate the work of the editorial team in facilitating the dissemination of innovative approaches such as the Bonori Method.” Best regards, Dr. Giselle Pentón-Rol.
