AUCTORES
Globalize your Research
Research Article | DOI: https://doi.org/10.31579/2690-1919/459
1 Department of Internal Medicine, National Taiwan University Hospital, Hsin-Chu Branch, Taiwan.
2 Department of Food Science, National Chiayi University, Chiayi, Taiwan.
3 Department of Biochemical Science and Technology, National Chiayi University, Chiayi, Taiwan.
# Qiao-Zhen Chang, and Bo-Cheng Mu contributed equally.
*Corresponding Author: Yun-Wei Lin, Consultant Division of pediatric orthopedics, Department of Biochemical Science and Technology, National Chiayi University, Chiayi, Taiwan.
Citation: Jen-Chung Koa, Jyh-Cheng Chenb, Ching-Hsiu Huangb, Pei-Jung Chenc, Yun-Wei Linc, et al., (2025), Epigallocatechin-3-Gallate Potentiates the Cytotoxic Effect of Erlotinib in Non-Small-Cell Lung Cancer Cells by P38 Mapk Mediated Xrcc3 Downregulation, J Clinical Research and Reports, 18(1); DOI:10.31579/2690-1919/459
Copyright: © 2025, Yun-Wei Lin. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received: 11 December 2024 | Accepted: 06 January 2025 | Published: 16 January 2025
Keywords: epigallocatechin gallate; xrcc3; erlotinib; cytotoxicity; non-small-cell lung cancer
Green tea polyphenols, particularly (-)-epigallocatechin gallate (EGCG), are thought to be responsible for the major physiologic benefits. XRCC3 participates in homologous recombination (HR) to repair DNA double-strand breaks (DSBs). The use of EGCG in combination with erlotinib to increase survival in patients with non-small-cell lung cancer (NSCLC) is still debatable and requires further research.
Materials and Methods
We used the MTS, trypan blue dye exclusion and colony-formation ability assay to determine whether EGCG alone or in combination with erlotinib had cytotoxic effects on the four NSCLC cell lines. Real-time PCR was conducted to measure the amounts of XRCC3 mRNA. The protein levels of phosphorylated p38 MAPK and XRCC3 were determined by Western blot analysis.
Results
We demonstrated that EGCG can amplify erlotinib cytotoxicity in NSCLC cells. EGCG treatment promote p38 MAPK activation and decreased XRCC3 mRNA and protein levels; moreover, combined treatment with erlotinib resulted in a significant reduction in XRCC3 protein levels and triggered XRCC3 degradation via a 26S proteasome-dependent pathway. The p38 MAPK activity was greatly increased by constitutively active MKK6 vector expression (MKK6E) and could promote XRCC3 degradation, which enhanced erlotinib and EGCG cytotoxicity. Conversely, SB203580, a p38 MAPK inhibitor, could prevent p38 MAPK activation and restore XRCC3 levels that had been decreased by erlotinib and improve cell survival.
Conclusion
Overall, our findings imply that XRCC3 downregulation is one of the primary mechanisms boosting the lethal effects of erlotinib by EGCG in NSCLC cells.
When single-stranded DNA (ssDNA) invades a homologous duplex DNA, which is fueled by the RAD51/RecA family of ATPases, homologous recombination (HR) pathways, are involved in the repair of DNA double-strand breaks (DSBs). Rad51 is a key component of HR pathways; ssDNA and RAD51 combine to create a nucleoprotein filament that performs homology search and catalyzes strand exchange [1]. To induce stable filament assembly and/or nucleate Rad51 filament formation on ssDNA in vivo, numerous variables are needed. RAD51 paralogs, which only slightly resemble RAD51 in the ATPase domain, are one of the elements contributing to this process. RAD51B [2], RAD51C [3], RAD51D [4], or complementation of radiation-sensitive Chinese hamster cell mutants XRCC2 [5] and XRCC3 [6] are the five canonical Rad51 paralogs in mammalian cells. The paralogs have been demonstrated to form two complexes biochemically, i.e., RAD51C–XRCC3 and RAD51B–RAD51C–RAD51D–XRCC2 [7]. At DSB sites, XRCC3 encourages Rad51 assembly [8]. To create a D-loop, the resulting RAD51-ssDNA filaments invade other unharmed homologous sequences. Eventually, gene conversion is the outcome of DNA synthesis from the invasive strand [9].
Non-small-cell lung cancer (NSCLC), which makes up roughly 85% of all lung cancer cases, has the highest mortality rate worldwide [10]. A shorter survival and poor prognosis are linked to tyrosine kinases (TKs) of the epidermal growth factor receptor (EGFR) or their ligands in NSCLC [11]. To prevent EGFR activation, small compounds known as TK inhibitors, such as erlotinib, can block the ATP-binding site on the receptor’s catalytic domain [12].
Green tea is made from the leaves of Camellia sinensis, a member of the Theaceae family, and exerts positive effects on human health, including anticancer, anti-obesity, antidiabetic, anticardiovascular, anti-infectious, and anti-neurodegenerative properties [13]. The majority of these biological effects are thought to be caused by (-)-epigallocatechin gallate (EGCG), the most prevalent catechin in green tea and the subject of earlier research. Moreover, 240–320 mg of catechins, of which 60%–65% is made up of EGCG, are present in a cup of green tea that is generally brewed using 2.5 g of tea leaves [14]. In the study by Wang et al., the extent of colon cancers in rats receiving dimethylhydrazine was reduced by EGCG [15]. EGCG, a well-known antioxidant, inhibits reactive oxygen species (ROS), which encourages oxidative DNA damage, mutagenesis, and tumor development and has anticancer effects [16], because EGCG-induced ROS production can activate AMPK by upregulating Ca2+/calmodulin-dependent protein kinase (CaMKK) and/or liver kinase B1 (LKB1) [17, 18], which inhibits the mechanistic target of rapamycin kinase (mTOR), having anticancer effects.
EGCG lessens side effects and strengthens anticancer effects; thus, it has been shown to have an additive or synergistic effect with chemopreventive medicines [19]. In previous study, EGCG inhibits cell proliferation and migration in NSCLC harboring wild-type EGFR (A549 cell line), in-frame deletion of exon 19 mutation (HCC827 cell line), and L858R/T790M double mutation (H1975 cell line) [20]. Moreover, the co-administration of EGCG and EGFR TK inhibitor gefitinib synergistic suppresses the tumor growth in the A549 xenograft mouse model [21]. In this study, we describe how EGCG affects XRCC3 expression and EGCG cytotoxicity in NSCLC cells. We demonstrate that erlotinib and EGCG combined therapy can lower XRCC3 protein levels in lung cancer cells by activating p38 MAPK. EGCG cytotoxicity is enhanced when the p38 MAPK activity is amplified and XRCC3 expression is downregulated. By modifying the XRCC3 and p38 MAPK signaling pathways, this study may offer a novel and more effective therapeutic approach to lung cancer treatment.
Chemicals and antibodies
EGCG was obtained from Sigma Chemical (St. Louis, MO, USA). MG132, SB203580, and N-acetyl-Leu-Leu-norleucinal (ALLN) were provided by Calbiochem-Novabiochem (San Diego, CA, USA). Phospho-p38 MAPK (Thr180/Tyr182) and phospho-MKK3 (Ser189)/MKK6 (Ser207) antibodies were purchased from Cell Signaling (Beverly, MA, USA). Abcam (Santa Cruz, CA) provided the rabbit polyclonal antibodies against XRCC3 (ab103070). Santa Cruz Biotechnology (TX, USA) provided the antibodies against p38 (C-20) (sc-535), MKK3 (N-20) (sc-959), and Actin (I-19) (sc-1616). Abcam provided the antibodies against XRCC3 (ab103070).
Cell lines and culture
RPMI-1640 complete medium supplemented with sodium bicarbonate (2.2% w/v), l-glutamine (0.03% w/v), penicillin (100 units/mL), streptomycin (100 µg/mL), and fetal calf serum (10%) was used to culture the human lung carcinoma cells H520, H1703, A549, and H1975, which were obtained from the American Type Culture Collection (Manassas, VA, USA).
Western blot analysis
After different treatments, the cells were rinsed twice with cold phosphate-buffered saline (PBS) solution and lysed in whole-cell extract buffer (20 mM HEPES [pH 7.6]), 75 mM NaCl, 2.5 mM MgCl2, 0.1 mM EDTA, 0.1% Triton X-100, 0.1 mM Na3VO4, 50 mM NaF, 1 µg/mL leupeptin, 1 µg/mL aprotinin, 1 µg/mL pepstatin, and 1 mM 4-(2- Equal amounts of proteins from each set of experiments were subjected to Western blot analysis as previously described [22].
Transfection with small-interfering RNA and MKK6E vectors
Sense-strand sequences for XRCC3 (s14948) siRNA duplexes were purchased from Thermo Fisher Scientific. By using Lipofectamine 2000 (Invitrogen), siRNA duplexes (200 nM) were transfected into cells for 24 h. The transfection of plasmids was accomplished as previously mentioned. Exponentially expanding human lung cancer cells (106) were plated for 18 h before Lipofectamine was used to transfect NSCLC cells with MKK6E (a constitutively active version of MKK6), as previously described [23].
Quantitative real-time polymerase chain reaction (PCR)
PCR tests were carried out on an ABI Prism 7900HT, as directed by the manufacturer. The SYBR Green PCR Master Mix (Applied Biosystems, MA, USA) was used to detect the amplification of particular PCR products. Data were standardized to the housekeeping gene glyceraldehyde 3-phosphate dehydrogenase (gapdh) for each sample. XRCC3 forward primer, 5'- AACCCGCGGGAGAGTCCCCA -3'; XRCC3 reverse primer, 5'- AAAGCCTGTGGGAGGCCCGA -3'; gapdh forward primer, 5'- CATGAGAAGTATGACAACAGCCT -3'; and gapdh reverse primer, 5'- AGTCCTTCCACGATACCAAAGT -3' were used in this study. Analysis was performed using the comparative CT value approach. The results were standardized to the housekeeping gene Gapdh for each sample.
Cell viability assay
An in vitro 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenol)-2- (4-sulfophenyl) Tetrazolium (MTS) assay was conducted. On 96-well tissue culture plates, 5000 cells were cultivated for each well. Drugs were introduced after plating to measure cell viability. After the culture period, 20 µL of the MTS solution (CellTiter 96 Aqueous One Solution Cell Proliferation Assay; Promega, Madison, WI, USA) was added. The cells were then cultured for another 2 h, and the absorbance was measured at 490 nm using an ELISA plate reader (Biorad Technologies, Hercules, CA, USA).
Combination index analysis of drug interactions
The effects of a 1:1 or 2:1 mixture of EGCG and erlotinib were examined using the MTS assay. To evaluate if a combination is synergistic (combination index [CI] < 0 xss=removed> 1.1), the CI, which considers the complete shape of the cell viability curve, was calculated using the computer program Calcusyn (Biosoft, Oxford, UK). The means of the CI values at fractions affected (FA) of 0.90, 0.75, and 0.50 were used to determine the difference between the three independent experiments.
Trypan blue dye exclusion assay
Erlotinib and/or EGCG were given to the cells for 24 h. Trypan blue dye cannot penetrate living cells, but it can enter dead ones. The number of trypan blue-stained cells and the percentage of dead cells were counted using a hemocytometer.
Colony-forming ability assay
Cells were trypsinized to count the number of cells right after the drug treatment and then washed with PBS. For each treatment, the cells were seeded in triplicate at a density of 500–1000 cells per 60 mm Petri dish. The cell colonies were stained with 1% crystal violet solution in 30% ethanol after the cells had grown for 10–14 days. Cytotoxicity was calculated by dividing the number of colonies in the treated cells by the number of colonies in the untreated control.
Experiments were conducted three or four times for each protocol. The results are expressed as the mean and the standard error of the mean (SEM). SigmaPlot 14.0 was used (Systat Software, San Jose, CA, USA). IBM SPSS Statistics for Windows version 20.0 (IBM Corp., Armonk, NY, USA) was used to examine the data. To see how treatments and time affected changes in cell viability, one-way analysis of variance was used to analyze differences between the two groups. This was followed by a Bonferroni post hoc test to ensure that all variables had a normal distribution. Differences were deemed significant at P < 0>
EGCG and erlotinib inhibited cell survival in NSCLC cells
We used the MTS assay to first determine whether EGCG alone or in combination with erlotinib had cytotoxic effects on the four NSCLC cell lines H520 (wild-type EGFR), H1703 (wild-type EGFR), A549 (wild-type EGFR), and H1975 (L858R/T790M). NSCLC cells dose- and time-dependently responded to EGCG, as depicted in Figure. 1A. In addition, the combined therapy of erlotinib (0.5–10 mM) and EGCG (0.5–20 mM) promote lung cancer cell cytotoxicity than either drug used alone (Figure. 1B). The CI was determined to ascertain the type of additive effect from EGCG and erlotinib (Table 1).
| Drug combination | CI Values Calculated for Different Effects | ||||||
| Cell line | (erlotinib:EGCG) | ED50 | ED75 | ED90 | |||
| H520 | (1:1) | 0.70 | ±0.03 | 1.51 | ±0.08 | 3.27 | ±0.42 |
| (1:2) | 0.74 | ±0.03 | 1.76 | ±0.22 | 4.67 | ±1.23 | |
| H1703 | (1:1) | 0.65 | ±0.07 | 1.01 | ±0.16 | 1.59 | ±0.40 |
| (1:2) | 0.29 | ±0.02 | 0.30 | ±0.01 | 0.34 | ±0.01 | |
| A549 | (1:1) | 0.35 | ±0.03 | 0.37 | ±0.07 | 0.40 | ±0.12 |
| (1:2) | 0.63 | ±0.16 | 1.57 | ±0.46 | 3.93 | ±1.32 | |
| H1975 | (1:1) | 0.50 | ±0.06 | 0.96 | ±0.08 | 1.85 | ±0.20 |
| (1:2) | 0.14 | ±0.02 | 0.20 | ±0.02 | 0.28 | ±0.03 | |
Data represent meanCI calculated fromthree independent experiments ±standard deviations | |||||||
| CI < 0.9 indicates synergism, CI = 0.9-1.1indicates additivity, and CI > 1.1 antagonism. | |||||||
Table 1: Values of combination index (CI) for erlotinib-EGCG interactions at different effects: ED50, ED75 and ED90.
The interactions between EGCG and erlotinib were primarily additive and antagonistic at the effect levels represented as ED90 and ED75; however, at ED50, they proved to be synergistic (CI < 0.1). On the evaluation of NSCLC cell death using the trypan blue exclusion assay, EGCG and erlotinib combined therapy had an additive effect on cell death (Figure. 1C). Moreover, erlotinib and EGCG combined therapy affected cell survival for up to 3 days in NSCLC cell lines (Figure. 1D). In these four NSCLC cells, the EGCG and erlotinib combined therapy inhibited cell growth more than erlotinib alone (Figure. 1D). The colony-formation assay was then used to determine if erlotinib and/or EGCG affect longterm clonogenic cell survival. In H520 (22.57% vs. 74.00%), H1703 (45.73% vs. 73.53%), A549 (45.97% vs. 70.83%), and H1975 (30.23% vs. 63.13%) cells, the colony-forming ability of EGCG and erlotinib combined treatment was lower than that of erlotinib treatment (Figure. 2).

Figure 1: Figure 1: Treatment of EGCG alone or combination with erlotinib response to cell survival in NSCLC cells. (A) For 24, 48, and 72 h, cells were exposed to EGCG at different concentrations (2.5, 5, 10, 20, 40, 60, 80, and 100 µM). The MTS assay was used to measure cell viability. (B) EGCG enhanced erlotinib cytotoxicity in NSCLC cells. The MTS assay was used to determine the cell viability after EGCG and erlotinib combined therapy at a ratio of 1:1 (upper panel) or 1:2 (lower panel). Three separate trials were used to gather the results (mean ± SEM). Using the one-way ANOVA to compare cells treated with erlotinib alone vs. EGCG/erlotinib combination, [a] ** signifies p < 0.01. Using the one-way ANOVA to evaluate cells treated with EGCG alone against EGCG and erlotinib combination, [b] ** p <0.01. (C) Unattached and attached cells were collected and stained with trypan blue after cells had been exposed to various dosages of EGCG (10 µM)/erlotinib for 24 h, and dead cells were manually counted. The population of dead cells was indicated by the proportion of trypan blue-positive cells, and the standard error was computed from three separate experiments. Using one-way ANOVA, the difference between cells treated with erlotinib alone vs. the EGCG/erlotinib combination, **p <0.001, *p < 0.05. (D) The number of living cells was counted using the MTS assay after the cells had been exposed to EGCG (20 µM) and erlotinib (10 µM) for 24, 48, and 72 h. Three separate trials were used to generate the results (mean ± SEM). Comparing cells treated with EGCG, erlotinib, or their combination revealed **p <0.01, using Student’s t-test.
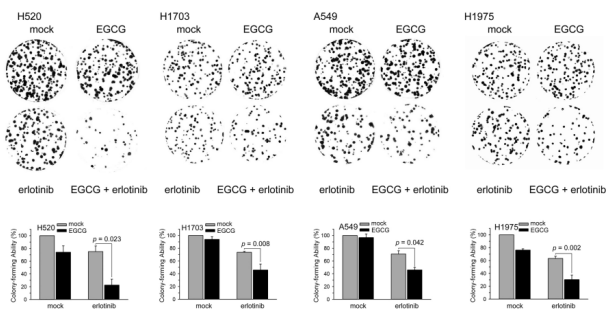
Figure 2: EGCG reduced erlotinib-induced colony-formation ability in NSCLC cells. After being exposed to EGCG (20 µM) and/or erlotinib (10 µM) for 24 h, cells were tested for their capacity to form colonies using the colony-formation assay. Three separate trials were used to gather the results (mean ± SEM). ** p < 0.01 when comparing cells treated with erlotinib alone against EGCG/erlotinib combination using a one-way ANOVA.
EGCG and erlotinib combined therapy additively decreased cellular XRCC3 protein levels
To examine the molecular mechanisms of EGCG in enhancing the cytotoxic effect of erlotinib on human lung cancer cell lines, NSCLC cells were exposed to various concentrations of EGCG alone or erlotinib and EGCG combination for 24 h. The protein levels of phosphorylated p38 MAPK and XRCC3 were then determined by Western blot analysis. EGCG dramatically decreased the levels of XRCC3 protein but increased the levels of phosphorylated p38 MAPK (Figure. 3A). In addition, EGCG and erlotinib combined treatment reduced XRCC3 levels in NSCLC cells (Figure. 3C).
EGCG decreased XRCC3 mRNA levels in erlotinib-treated NSCLC cells
EGCG and erlotinib combined treatment was given to NSCLC cells for 24 h to determine if the treatment-induced reduction in XRCC3 protein levels occurred at the transcriptional level. Real-time PCR was conducted to measure the amounts of XRCC3 mRNA in total RNA. XRCC3 mRNA levels reduced by EGCG was dose-dependent manner (Figure. 3B). In 4 different NSCLC cell lines, EGCG and erlotinib combined treatment had a more suppressive effect on XRCC3 mRNA levels (Figure. 3D). Next, we investigated potential mechanisms for posttranscriptional regulation of XRCC3 transcripts under EGCG and/or erlotinib treatment. In the presence of actinomycin D (an inhibitor of RNA synthesis), EGCG and erlotinib combined treatment showed less XRCC3 mRNA stability than the erlotinib treated alone in H520, H1703, and H1975, but not in A549 cells (Figure. 3E; Table 2).

Figure 3: In human NSCLC cell lines, erlotinib and/or EGCG decreased XRCC3 mRNA and protein levels. (A, B) NSCLC cells (106 ) were grown in full media for 18 h before being treated with various EGCG doses for 24 h. Following treatment, total RNA was extracted and submitted to real-time PCR for examination of XRCC3 mRNA expression and Western blot analysis for the quantification of the levels of XRCC3, phospho-MKK3/6, phospho-p38 MAPK, actin, MKK6, and p38 MAPK proteins. (C) Erlotinib (5 and 10 µM) and/or EGCG (20 µM) were administered to NSCLC cells (106 ) after they had been grown in full medium for 18 h. The treatment lasted for 24 h. Following treatment, cell extracts were utilized to evaluate the levels of XRCC3 protein after the aforementioned treatment. Four investigations are represented by Western blots. (D) Cell extracts were utilized to evaluate the amounts of XRCC3 mRNA after the aforementioned treatment. Total RNA was extracted, and real-time PCR was used to ascertain XRCC3 mRNA expression. Four separate experiments were conducted, and the findings (means standard deviation) were collected from each. Using the oneway ANOVA to compare cells treated with EGCG and erlotinib separately or together, ** p < 0.01. (E) NSCLC cells were treated with EGCG (20 µM) and/or erlotinib (10 µM) for 24 hours in the presence or absence of actinomycin D (1 µg/mL) for 3, 6, or 9 hours. The total RNA was isolated, and XRCC3 mRNA expression levels were determined using real-time PCR. The results (mean ± SEM) were acquired from three independent experiments. (F) Treatment with erlotinib plus EGCG causes XRCC3 to be proteolyzed by 26S proteasome. EGCG (20 µM) and erlotinib (10 µM) were administered to NSCLC cells for 18 h, followed by a 6-h co-treatment with the 26S proteasome inhibitors MG132 (10 µM) or ALLN (10 µM). For Western blotting, entire cell extracts were collected. Four investigations are represented by Western blots.
| XRCC3 mRNA half-life (h) | ||||
| cells | control | EGCG | erlotinib | EGCG+erlotinib |
| H520 | 7.89 | 6.19 | 6.98 | 5.34 |
| H1703 | 8.71 | 5.57 | 7.13 | 3.87 |
| A549 | 3.96 | 3.34 | 3.50 | 2.81 |
| H1975 | 8.71 | 4.99 | 7.04 | 3.92 |
Table 2: XRCC3 mRNA half-life (h) for EGCG and/or erlotinib treatment.
EGCG and erlotinib combined treatment decreased XRCC3 protein levels via 26S proteasome-directed proteolysis
To determine if the 26S proteasome was responsible for the degradation of the XRCC3 protein, MG132, or ALLN, two 26S proteasome inhibitors were co-added to NSCLC cells that had been exposed to erlotinib and EGCG. Consequently, MG132 and ALLN stopped the XRCC3 protein degradation induced by EGCG and erlotinib combined treatment (Figure. 3F). Together, EGCG and erlotinib combined treatment caused XRCC3 protein degradation through the 26S proteasome-dependent pathway, which was the mechanism for reducing XRCC3 protein levels.
Influence of the p38 MAPK signaling pathway on XRCC3 protein levels in lung cancer cells exposed to EGCG and erlotinib combined treatment
To evaluate the role of p38 MAPK activation in regulating XRCC3 expression, we investigated whether blocking p38 MAPK activity could
affect XRCC3 expression in NSCLC cells. NSCLC cells cultured with a p38 MAPK inhibitor (SB203580) result in high XRCC3 mRNA and protein levels (Figure. 4A, 4B). As can be seen in Figure. 4C and 4D, SB203580 reversed the EGCG-induced decrease in XRCC3 mRNA and protein levels in NSCLC cells.
In addition, NSCLC cells were transiently transfected with plasmids containing MKK6E (a constitutively active form of MKK6, which is the upstream protein kinase of p38 MAPK) to determine whether the p38 MAPK signaling pathway was involved in the regulation of XRCC3 protein levels. In NSCLC cells, transfection of MKK6E could promote p38 MAPK activation while lowering XRCC3 mRNA and protein levels (Figure. 4E, 4F). In addition, MKK6E expression dramatically reduced XRCC3 mRNA and protein levels in EGCG-treated NSCLC cells (Fig. 4E, 4F). Our findings suggest that MKK6-p38 MAPK is the upstream signal for reducing XRCC3 protein levels in NSCLC cells.

Figure 4.p38 MAPK activation downregulates XRCC3 mRNA and protein expression in EGCG-treated lung cancer cells. (A, B) NSCLC cells were treated for p38 MAPK inhibitor SB203580 (2.5, 5, and 10 µM) for 24 h. (C, D) NSCLC cells were pretreated for 1 h with the p38 MAPK inhibitor SB203580 (5 µM) before being co-treated for 24 h with EGCG (20 µM). Following treatment, the levels of XRCC3 protein and mRNA in the cell extracts were determined by Western blot analysis and real-time PCR, respectively. (E, F) MKK6E expression vectors were transfected into NSCLC cells. Once the cells had been expressed for 24 h, they were treated with EGCG (40 and 60 M) for 24 h. Total RNA was then extracted, and XRCC3 mRNA expression was determined by real-time PCR (E). Using Student’s t-test, the EGCG-treated cells transfected with the pcDNA3 or MKK6E vectors were compared, **p < 0.01. For the comparison of cells transfected with pcDNA3 or MKK6E vectors, #p < 0.01, using one-way ANOVA. (F) Western blot analysis was used to check the cell extracts after the aforementioned treatment.
p38 mapk activation and downregulation of xrcc3 expression enhanced egcg-induced cytotoxic effects
To investigate the role of p38 MAPK activation in the cytotoxic effect of EGCG, cells were transfected with pcDNA3 or MKK6E expression vectors and then treated with EGCG for 24 h. MKK6E transfection enhanced the cytotoxic and growth-inhibitory effects of EGCG as a means of examining the role of p38 MAPK activation in the cytotoxic effect of EGCG (Fig. 5A, 5B). By contrast, blocking the p38 MAPK activity by SB2023580 could reverse the cytotoxicity and growth-inhibitory effect of EGCG (Fig. 5C, 5D). Furthermore, by using certain siRNA duplexes, XRCC3 mRNA expression was knocked down to ascertain the role of XRCC3 in the cytotoxic effect caused by EGCG. The MTS assay was used to quantify the cytotoxicity induced by EGCG and si-XRCC3 RNA transfection. In Fig. 5E, si-XRCC3 RNA transfection increased the cytotoxicity induced by EGCG. Taken together, both p38 MAPK activation and XRCC3 expression reduction could enhance EGCG-induced cytotoxicity and growth inhibition in NSCLC cells.

Figure 5. p38 MAPK activation and XRCC3 downregulation enhanced EGCG cytotoxicity in NSCLC cell lines. (A) EGCG was applied to NSCLC cells for 24 h after MKK6E (5 µg) or pcDNA3 (5 µg) expression vector transfection. The MTS assay was used to evaluate cytotoxicity. The results (mean ± SEM) were acquired from three independent experiments. (B) After being transfected with pcDNA3 or MKK6E expression vector, the cells were treated with EGCG (20 µM) for 24, 48, and 72 h, and the living cells were counted using the MTS assay. The results (mean ± SEM) were acquired from three independent experiments. ** p < 0>< 0.01, *p < 0.05 (one-way ANOVA) was used. (D) Cells were treated with SB203580 (10 µM) and/or EGCG (40 µM) for 1–3 days, and the living cells were then counted using the MTS assay. The results (mean ± SEM) were acquired from four independent experiments. ** p < 0.01 using the one-way ANOVA to compare cells treated with EGCG alone or GCG and SB203580 combination. (E) The cells were treated with EGCG (20 µM) for 1–4 days following transfection with either XRCC3 siRNA or scrambled siRNA. Living cells were then identified using the MTS assay. Three separate trials were used to generate the results (mean ± SEM). Using Student’s t-test, EGCG-treated cells transfected with either XRCC3 siRNA or scrambled siRNA were compared; **p < 0.01.
Role of XRCC3 and p38 MAPK in EGCG-enhancing erlotinib-inducing cytotoxicity in lung cancer cells
To assess the effect of XRCC3 downregulation on erlotinib cytotoxicity, in erlotinib-treated NSCLC cells, transfection of the si-XRCC3 RNA duplex could drastically reduce XRCC3 mRNA levels (Fig. 6A). In addition, lung cancer cells were treated with EGCG or transfected with si-XRCC3 RNA to suppress XRCC3 expression. EGCG and si-XRCC3 RNA co-treatment could enhance erlotinib cytotoxicity (Fig. 6B). In addition, a p38 MAPK inhibitor was used as pretreatment for NSCLC cells before erlotinib and EGCG administration. The MTS assay was used to measure cell survival. As shown in Fig. 6C, SB203580 could enhance the cell survival that was previously suppressed by EGCG and/or erlotinib. By contrast, the transfection of MKK6E vectors could further reduce cell viability, which is consistent with the cytotoxic effect of EGCG and erlotinib, which was caused by p38 MAPK activation (Fig. 6D). Therefore, EGCG-enhancing erlotinib-inducing cytotoxicity through downregulating XRCC3 expression and activated p38 MAPK in NSCLC cells.

Figure 6. The viability of the EGCG- and erlotinib-induced cells is recovered by inhibiting p38 MAPK activity. (A) Before being treated with erlotinib in complete medium for 24 h, NSCLC cells were transfected with siRNA duplexes (200 nM) targeted to XRCC3 or scrambled (control) in a complete medium. After treatment, cell extracts were used to determine XRCC3 mRNA and protein using real-time PCR. (B) Using lipofectamine, the cells were transfected to express si-XRCC3 RNA. Following 24 h of expression, the cells were treated with erlotinib or EGCG (20 µM) for an additional 24 h, and the MTS assay was used to assess cytotoxicity. c**p < 0.01, using one-way ANOVA for comparison of the erlotinib-treated cells that were transfected with si-XRCC3 or scrambled siRNA. d**p < 0.01, using one-way ANOVA for comparison of the erlotinib-treated or erlotinib and EGCGco-treated cells. (C) NSCLC cells were pretreated with SB2023580 for 1 h and then co-treated with EGCG (10 µM) and erlotinib (10 µM) for 24 h. Cytotoxicity was assessed using the MTS assay. **p < 0.01 using the one-way ANOVA to compare cells treated with EGCG and erlotinib in SB2023580 or DMSO vector-transfected cells. (D) After being transfected with pcDNA3 or MKK6E expression vector, the cells were treated with EGCG and erlotinib for 24 h, and living cells were counted using the MTS assay. The results (mean ± SEM) were acquired from three independent experiments. ** p < 0.01 using the one-way ANOVA to compare cells treated with EGCG and erlotinib in MKK6E or pcDNA3-transfected cells.
Cancer is the world’s most serious challenge for public health in terms of morbidity and mortality. This pathogenesis is caused by several factors, including modifications in several biological processes and cell signaling pathways. Moreover, natural substances or active components of herbs contribute significantly to the improvement of the effectiveness of anticancer treatments and lessen their toxicity. It has been anticipated that pharmacological combinations made of dietary supplements and natural items will produce the same results as traditional chemotherapy with fewer side effects [24]. In this study, EGCG may promote erlotinib cytotoxicity in NSCLC cells, which is linked to a reduction in XRCC3 expression.
Lung cancer, melanoma, breast cancer, leukemia, and colon cancer are a few of the human cancer cell lines that have shown growth suppression and apoptosis when exposed to EGCG in vitro [25, 26]. Lower levels of phospho-c-jun and phospho-ERK1/2 in lung adenomas and carcinomas, decreased cell proliferation, increased apoptosis, and decreased angiogenesis were all linked to the prevention of lung tumor by EGCG in A/J mice [27]. By increasing p53 expression, EGCG is a key factor in the suppression of anchorage-independent development of human lung cancer cells. Moreover, EGCG activity can significantly boost p53 stability and promote p53 nuclear localization [28]. By inhibiting HO-1 expression and enhancing the antitumor effects of EGCG, metformin also amplified EGCG-induced ROS production, which ultimately led to apoptosis [29]. Serum samples from patients with cancer had lower hsa-mir-485-5p levels. Meanwhile, EGCG contributed to the upregulation of hsa-mir-485-5p expression, and hsa-mir-485-5p imitators significantly enhanced cell apoptosis and reduced cancer cell proliferation [30]. Cleaved caspase-3 and Bax were further expressed when EGCG-induced apoptosis was applied to A549 lung cancer cells; however, Bcl-xL was less expressed [31]. Moreover, EGCG effectively reduced lung cancer stem cell activity by lowering lung cancer stem cell markers, preventing the development of tumorspheres, reducing proliferation, and promoting apoptosis [32]. The ability of EGCG to increase interleukin-6 production was explained by its cytotoxic effects on both parental lung cancer cells and their cisplatin-resistant counterparts. However, EGCG did not affect the downstream effector signal transducers of interleukin-6 and activators of transcriptional 3 phosphorylation [33].
The combination of EGCG and curcumin can inhibit tumor growth without causing any serious side effects in the lung cancer xenograft nude mice model [34]. Furthermore, combination of erlotinib and EGCG with classical chemotherapeutics methotrexate or actinomycin-D lead the human placenta choriocarcinoma cells to apoptosis [35]. Our study demonstrated that the combination of EGCG and erlotinib has synergistic cytotoxicity in NSCLC cells through p38 MAPK-mediated XRCC3 downregulation. Taken together, EGCG presents a synergistic effect when co-treated with chemotherapy medicines in different types of tumor cell lines.
Cell apoptosis, differentiation, and death are a few of the crucial physiological processes in which MAP kinases are involved. The p38 MAP kinases, ERK, and c-JNK are the three main MAP kinases found in mammalian cells [25]. Studies have investigated the established role of MAP kinase pathways in EGCG’s control of AP-1 activity. In Ha-ras-transformed human bronchial cells, EGCG treatment has been demonstrated to decrease the phosphorylation c-Jun, ERK1/2, ELK1, and MEK1/2 [36]. Contrary to these reports, EGCG has been shown to significantly boost AP-1 factor-associated responses via a MAP kinase signaling mechanism in normal human keratinocytes, indicating that the signaling mechanism of EGCG activity may differ noticeably in various cell types [37]. In mammalian cells, the JNK and p38 MAPK pathways are parallel MAP kinase cascades [38]. EGCG-induced modulation of MAP kinases may offer fresh approaches for the prevention or treatment of human cancer because the MAP kinase pathway is frequently dysregulated in several malignancies. In rat hepatoma, leukemia, and lung cancer cells, tea catechin EGCG reduces DNA synthesis [25, 27]. Lower levels of phospho-c-jun and phospho-ERK1/2 in lung adenomas and carcinomas, decreased cell proliferation, increased apoptosis, and decreased angiogenesis were all linked to the preventive effect of EGCG on A/J mouse lung tumor [27]. Moreover, the anti-aging gene Sirtuin 1 is important to lung function and the treatment of NSCLC. EGCG has been shown to regulate and inhibit Sirtuin 1 and Sirtuin 1 regulates p38 MAPK. EGCG and erlotinib combined therapy to inhibit XRCC3 expression in NSCLC as a new combined therapy may require the measurement of Sirtuin 1 in these experiments for the long-term survival of patients with NSCLC. Sirtuin 1 activators and inhibitors may determine the EGCG and erlotinib combined therapy [39-41].
To the best of our knowledge, this study is the first to show that EGCG reduces the levels of cellular XRCC3 protein and mRNA in NSCLC cells through the activation of MKK3/6-p38 MAPK. In addition, one of the main elements contributing to the enhancement of erlotinib’s cytotoxic effects is the EGCG-induced down-modulation of XRCC3 protein. As a result, EGCG and erlotinib combined therapy to inhibit XRCC3 expression in NSCLC may be a new combined therapy that can synergistically enhance cytotoxic effects. In vivo animal studies and human clinical trials are needed to confirm that this proposal is effective in enhancing the overall survival of patients with NSCLC without additive side effects.
Not applicable.
The authors declare that they have no conflict of interest.
We thank Dr. Chia-Che Chang and Dr. Jia-Ling Yang for providing us with expression plasmids for transfection.
This study protocol was reviewed and the need for approval was waived by Biosafety Committee and National Chiayi University. The immortalized cell lines used in this study were purchased from the American Type Culture Collection (Manassas, VA, USA). Ethical approval for use of these cells is not required in accordance with local/national guidelines. This study does not contain any studies with human participants or animals performed by any of the authors.
The authors declare that they have no conflict of interest.
This study was funded by grants from the Ministry of Science and Technology, Taiwan, Grant no. MOST 110-2314-B-002 -265 -MY3; MOST 110-2320-B-415-004, and the National Taiwan University Hospital, Hsin-Chu Branch, Taiwan.
Yun-Wei Lin, Jen-Chung Ko and Jyh-Cheng Chen contributed to conception, design, data analysis and interpretation, and drafting the manuscript. Ching-Hsiu Huang, Pei-Jung Chen, Qiao-Zhen Chang, Bo-Cheng Mu, Jou-Min Hsieh, Pei-Yu Tseng, Chen-Shan Chiang, Jun-Jie Chen and Yun-Wei Lin contributed to data acquisition, data interpretation, and critically revised the manuscript. All authors gave final approval and agreed to be accountable for all aspects of work ensuring integrity and accuracy.
All data generated or analysed during this study are included in this article. Further enquiries can be directed to the corresponding author.
Clearly Auctoresonline and particularly Psychology and Mental Health Care Journal is dedicated to improving health care services for individuals and populations. The editorial boards' ability to efficiently recognize and share the global importance of health literacy with a variety of stakeholders. Auctoresonline publishing platform can be used to facilitate of optimal client-based services and should be added to health care professionals' repertoire of evidence-based health care resources.

Journal of Clinical Cardiology and Cardiovascular Intervention The submission and review process was adequate. However I think that the publication total value should have been enlightened in early fases. Thank you for all.

Journal of Women Health Care and Issues By the present mail, I want to say thank to you and tour colleagues for facilitating my published article. Specially thank you for the peer review process, support from the editorial office. I appreciate positively the quality of your journal.
Journal of Clinical Research and Reports I would be very delighted to submit my testimonial regarding the reviewer board and the editorial office. The reviewer board were accurate and helpful regarding any modifications for my manuscript. And the editorial office were very helpful and supportive in contacting and monitoring with any update and offering help. It was my pleasure to contribute with your promising Journal and I am looking forward for more collaboration.

We would like to thank the Journal of Thoracic Disease and Cardiothoracic Surgery because of the services they provided us for our articles. The peer-review process was done in a very excellent time manner, and the opinions of the reviewers helped us to improve our manuscript further. The editorial office had an outstanding correspondence with us and guided us in many ways. During a hard time of the pandemic that is affecting every one of us tremendously, the editorial office helped us make everything easier for publishing scientific work. Hope for a more scientific relationship with your Journal.

The peer-review process which consisted high quality queries on the paper. I did answer six reviewers’ questions and comments before the paper was accepted. The support from the editorial office is excellent.

Journal of Neuroscience and Neurological Surgery. I had the experience of publishing a research article recently. The whole process was simple from submission to publication. The reviewers made specific and valuable recommendations and corrections that improved the quality of my publication. I strongly recommend this Journal.

Dr. Katarzyna Byczkowska My testimonial covering: "The peer review process is quick and effective. The support from the editorial office is very professional and friendly. Quality of the Clinical Cardiology and Cardiovascular Interventions is scientific and publishes ground-breaking research on cardiology that is useful for other professionals in the field.

Thank you most sincerely, with regard to the support you have given in relation to the reviewing process and the processing of my article entitled "Large Cell Neuroendocrine Carcinoma of The Prostate Gland: A Review and Update" for publication in your esteemed Journal, Journal of Cancer Research and Cellular Therapeutics". The editorial team has been very supportive.

Testimony of Journal of Clinical Otorhinolaryngology: work with your Reviews has been a educational and constructive experience. The editorial office were very helpful and supportive. It was a pleasure to contribute to your Journal.

Dr. Bernard Terkimbi Utoo, I am happy to publish my scientific work in Journal of Women Health Care and Issues (JWHCI). The manuscript submission was seamless and peer review process was top notch. I was amazed that 4 reviewers worked on the manuscript which made it a highly technical, standard and excellent quality paper. I appreciate the format and consideration for the APC as well as the speed of publication. It is my pleasure to continue with this scientific relationship with the esteem JWHCI.

This is an acknowledgment for peer reviewers, editorial board of Journal of Clinical Research and Reports. They show a lot of consideration for us as publishers for our research article “Evaluation of the different factors associated with side effects of COVID-19 vaccination on medical students, Mutah university, Al-Karak, Jordan”, in a very professional and easy way. This journal is one of outstanding medical journal.
Dear Hao Jiang, to Journal of Nutrition and Food Processing We greatly appreciate the efficient, professional and rapid processing of our paper by your team. If there is anything else we should do, please do not hesitate to let us know. On behalf of my co-authors, we would like to express our great appreciation to editor and reviewers.

As an author who has recently published in the journal "Brain and Neurological Disorders". I am delighted to provide a testimonial on the peer review process, editorial office support, and the overall quality of the journal. The peer review process at Brain and Neurological Disorders is rigorous and meticulous, ensuring that only high-quality, evidence-based research is published. The reviewers are experts in their fields, and their comments and suggestions were constructive and helped improve the quality of my manuscript. The review process was timely and efficient, with clear communication from the editorial office at each stage. The support from the editorial office was exceptional throughout the entire process. The editorial staff was responsive, professional, and always willing to help. They provided valuable guidance on formatting, structure, and ethical considerations, making the submission process seamless. Moreover, they kept me informed about the status of my manuscript and provided timely updates, which made the process less stressful. The journal Brain and Neurological Disorders is of the highest quality, with a strong focus on publishing cutting-edge research in the field of neurology. The articles published in this journal are well-researched, rigorously peer-reviewed, and written by experts in the field. The journal maintains high standards, ensuring that readers are provided with the most up-to-date and reliable information on brain and neurological disorders. In conclusion, I had a wonderful experience publishing in Brain and Neurological Disorders. The peer review process was thorough, the editorial office provided exceptional support, and the journal's quality is second to none. I would highly recommend this journal to any researcher working in the field of neurology and brain disorders.

Dear Agrippa Hilda, Journal of Neuroscience and Neurological Surgery, Editorial Coordinator, I trust this message finds you well. I want to extend my appreciation for considering my article for publication in your esteemed journal. I am pleased to provide a testimonial regarding the peer review process and the support received from your editorial office. The peer review process for my paper was carried out in a highly professional and thorough manner. The feedback and comments provided by the authors were constructive and very useful in improving the quality of the manuscript. This rigorous assessment process undoubtedly contributes to the high standards maintained by your journal.

International Journal of Clinical Case Reports and Reviews. I strongly recommend to consider submitting your work to this high-quality journal. The support and availability of the Editorial staff is outstanding and the review process was both efficient and rigorous.

Thank you very much for publishing my Research Article titled “Comparing Treatment Outcome Of Allergic Rhinitis Patients After Using Fluticasone Nasal Spray And Nasal Douching" in the Journal of Clinical Otorhinolaryngology. As Medical Professionals we are immensely benefited from study of various informative Articles and Papers published in this high quality Journal. I look forward to enriching my knowledge by regular study of the Journal and contribute my future work in the field of ENT through the Journal for use by the medical fraternity. The support from the Editorial office was excellent and very prompt. I also welcome the comments received from the readers of my Research Article.

Dear Erica Kelsey, Editorial Coordinator of Cancer Research and Cellular Therapeutics Our team is very satisfied with the processing of our paper by your journal. That was fast, efficient, rigorous, but without unnecessary complications. We appreciated the very short time between the submission of the paper and its publication on line on your site.

I am very glad to say that the peer review process is very successful and fast and support from the Editorial Office. Therefore, I would like to continue our scientific relationship for a long time. And I especially thank you for your kindly attention towards my article. Have a good day!

"We recently published an article entitled “Influence of beta-Cyclodextrins upon the Degradation of Carbofuran Derivatives under Alkaline Conditions" in the Journal of “Pesticides and Biofertilizers” to show that the cyclodextrins protect the carbamates increasing their half-life time in the presence of basic conditions This will be very helpful to understand carbofuran behaviour in the analytical, agro-environmental and food areas. We greatly appreciated the interaction with the editor and the editorial team; we were particularly well accompanied during the course of the revision process, since all various steps towards publication were short and without delay".

I would like to express my gratitude towards you process of article review and submission. I found this to be very fair and expedient. Your follow up has been excellent. I have many publications in national and international journal and your process has been one of the best so far. Keep up the great work.

We are grateful for this opportunity to provide a glowing recommendation to the Journal of Psychiatry and Psychotherapy. We found that the editorial team were very supportive, helpful, kept us abreast of timelines and over all very professional in nature. The peer review process was rigorous, efficient and constructive that really enhanced our article submission. The experience with this journal remains one of our best ever and we look forward to providing future submissions in the near future.

I am very pleased to serve as EBM of the journal, I hope many years of my experience in stem cells can help the journal from one way or another. As we know, stem cells hold great potential for regenerative medicine, which are mostly used to promote the repair response of diseased, dysfunctional or injured tissue using stem cells or their derivatives. I think Stem Cell Research and Therapeutics International is a great platform to publish and share the understanding towards the biology and translational or clinical application of stem cells.

I would like to give my testimony in the support I have got by the peer review process and to support the editorial office where they were of asset to support young author like me to be encouraged to publish their work in your respected journal and globalize and share knowledge across the globe. I really give my great gratitude to your journal and the peer review including the editorial office.

I am delighted to publish our manuscript entitled "A Perspective on Cocaine Induced Stroke - Its Mechanisms and Management" in the Journal of Neuroscience and Neurological Surgery. The peer review process, support from the editorial office, and quality of the journal are excellent. The manuscripts published are of high quality and of excellent scientific value. I recommend this journal very much to colleagues.

Dr.Tania Muñoz, My experience as researcher and author of a review article in The Journal Clinical Cardiology and Interventions has been very enriching and stimulating. The editorial team is excellent, performs its work with absolute responsibility and delivery. They are proactive, dynamic and receptive to all proposals. Supporting at all times the vast universe of authors who choose them as an option for publication. The team of review specialists, members of the editorial board, are brilliant professionals, with remarkable performance in medical research and scientific methodology. Together they form a frontline team that consolidates the JCCI as a magnificent option for the publication and review of high-level medical articles and broad collective interest. I am honored to be able to share my review article and open to receive all your comments.

“The peer review process of JPMHC is quick and effective. Authors are benefited by good and professional reviewers with huge experience in the field of psychology and mental health. The support from the editorial office is very professional. People to contact to are friendly and happy to help and assist any query authors might have. Quality of the Journal is scientific and publishes ground-breaking research on mental health that is useful for other professionals in the field”.

Dear editorial department: On behalf of our team, I hereby certify the reliability and superiority of the International Journal of Clinical Case Reports and Reviews in the peer review process, editorial support, and journal quality. Firstly, the peer review process of the International Journal of Clinical Case Reports and Reviews is rigorous, fair, transparent, fast, and of high quality. The editorial department invites experts from relevant fields as anonymous reviewers to review all submitted manuscripts. These experts have rich academic backgrounds and experience, and can accurately evaluate the academic quality, originality, and suitability of manuscripts. The editorial department is committed to ensuring the rigor of the peer review process, while also making every effort to ensure a fast review cycle to meet the needs of authors and the academic community. Secondly, the editorial team of the International Journal of Clinical Case Reports and Reviews is composed of a group of senior scholars and professionals with rich experience and professional knowledge in related fields. The editorial department is committed to assisting authors in improving their manuscripts, ensuring their academic accuracy, clarity, and completeness. Editors actively collaborate with authors, providing useful suggestions and feedback to promote the improvement and development of the manuscript. We believe that the support of the editorial department is one of the key factors in ensuring the quality of the journal. Finally, the International Journal of Clinical Case Reports and Reviews is renowned for its high- quality articles and strict academic standards. The editorial department is committed to publishing innovative and academically valuable research results to promote the development and progress of related fields. The International Journal of Clinical Case Reports and Reviews is reasonably priced and ensures excellent service and quality ratio, allowing authors to obtain high-level academic publishing opportunities in an affordable manner. I hereby solemnly declare that the International Journal of Clinical Case Reports and Reviews has a high level of credibility and superiority in terms of peer review process, editorial support, reasonable fees, and journal quality. Sincerely, Rui Tao.

Clinical Cardiology and Cardiovascular Interventions I testity the covering of the peer review process, support from the editorial office, and quality of the journal.

Clinical Cardiology and Cardiovascular Interventions, we deeply appreciate the interest shown in our work and its publication. It has been a true pleasure to collaborate with you. The peer review process, as well as the support provided by the editorial office, have been exceptional, and the quality of the journal is very high, which was a determining factor in our decision to publish with you.
The peer reviewers process is quick and effective, the supports from editorial office is excellent, the quality of journal is high. I would like to collabroate with Internatioanl journal of Clinical Case Reports and Reviews journal clinically in the future time.

Clinical Cardiology and Cardiovascular Interventions, I would like to express my sincerest gratitude for the trust placed in our team for the publication in your journal. It has been a true pleasure to collaborate with you on this project. I am pleased to inform you that both the peer review process and the attention from the editorial coordination have been excellent. Your team has worked with dedication and professionalism to ensure that your publication meets the highest standards of quality. We are confident that this collaboration will result in mutual success, and we are eager to see the fruits of this shared effort.

Dear Dr. Jessica Magne, Editorial Coordinator 0f Clinical Cardiology and Cardiovascular Interventions, I hope this message finds you well. I want to express my utmost gratitude for your excellent work and for the dedication and speed in the publication process of my article titled "Navigating Innovation: Qualitative Insights on Using Technology for Health Education in Acute Coronary Syndrome Patients." I am very satisfied with the peer review process, the support from the editorial office, and the quality of the journal. I hope we can maintain our scientific relationship in the long term.
Dear Monica Gissare, - Editorial Coordinator of Nutrition and Food Processing. ¨My testimony with you is truly professional, with a positive response regarding the follow-up of the article and its review, you took into account my qualities and the importance of the topic¨.

Dear Dr. Jessica Magne, Editorial Coordinator 0f Clinical Cardiology and Cardiovascular Interventions, The review process for the article “The Handling of Anti-aggregants and Anticoagulants in the Oncologic Heart Patient Submitted to Surgery” was extremely rigorous and detailed. From the initial submission to the final acceptance, the editorial team at the “Journal of Clinical Cardiology and Cardiovascular Interventions” demonstrated a high level of professionalism and dedication. The reviewers provided constructive and detailed feedback, which was essential for improving the quality of our work. Communication was always clear and efficient, ensuring that all our questions were promptly addressed. The quality of the “Journal of Clinical Cardiology and Cardiovascular Interventions” is undeniable. It is a peer-reviewed, open-access publication dedicated exclusively to disseminating high-quality research in the field of clinical cardiology and cardiovascular interventions. The journal's impact factor is currently under evaluation, and it is indexed in reputable databases, which further reinforces its credibility and relevance in the scientific field. I highly recommend this journal to researchers looking for a reputable platform to publish their studies.
Dear Editorial Coordinator of the Journal of Nutrition and Food Processing! "I would like to thank the Journal of Nutrition and Food Processing for including and publishing my article. The peer review process was very quick, movement and precise. The Editorial Board has done an extremely conscientious job with much help, valuable comments and advices. I find the journal very valuable from a professional point of view, thank you very much for allowing me to be part of it and I would like to participate in the future!”

Dealing with The Journal of Neurology and Neurological Surgery was very smooth and comprehensive. The office staff took time to address my needs and the response from editors and the office was prompt and fair. I certainly hope to publish with this journal again.Their professionalism is apparent and more than satisfactory. Susan Weiner

My Testimonial Covering as fellowing: Lin-Show Chin. The peer reviewers process is quick and effective, the supports from editorial office is excellent, the quality of journal is high. I would like to collabroate with Internatioanl journal of Clinical Case Reports and Reviews.

My experience publishing in Psychology and Mental Health Care was exceptional. The peer review process was rigorous and constructive, with reviewers providing valuable insights that helped enhance the quality of our work. The editorial team was highly supportive and responsive, making the submission process smooth and efficient. The journal's commitment to high standards and academic rigor makes it a respected platform for quality research. I am grateful for the opportunity to publish in such a reputable journal.
My experience publishing in International Journal of Clinical Case Reports and Reviews was exceptional. I Come forth to Provide a Testimonial Covering the Peer Review Process and the editorial office for the Professional and Impartial Evaluation of the Manuscript.

I would like to offer my testimony in the support. I have received through the peer review process and support the editorial office where they are to support young authors like me, encourage them to publish their work in your esteemed journals, and globalize and share knowledge globally. I really appreciate your journal, peer review, and editorial office.
Dear Agrippa Hilda- Editorial Coordinator of Journal of Neuroscience and Neurological Surgery, "The peer review process was very quick and of high quality, which can also be seen in the articles in the journal. The collaboration with the editorial office was very good."

I would like to express my sincere gratitude for the support and efficiency provided by the editorial office throughout the publication process of my article, “Delayed Vulvar Metastases from Rectal Carcinoma: A Case Report.” I greatly appreciate the assistance and guidance I received from your team, which made the entire process smooth and efficient. The peer review process was thorough and constructive, contributing to the overall quality of the final article. I am very grateful for the high level of professionalism and commitment shown by the editorial staff, and I look forward to maintaining a long-term collaboration with the International Journal of Clinical Case Reports and Reviews.
To Dear Erin Aust, I would like to express my heartfelt appreciation for the opportunity to have my work published in this esteemed journal. The entire publication process was smooth and well-organized, and I am extremely satisfied with the final result. The Editorial Team demonstrated the utmost professionalism, providing prompt and insightful feedback throughout the review process. Their clear communication and constructive suggestions were invaluable in enhancing my manuscript, and their meticulous attention to detail and dedication to quality are truly commendable. Additionally, the support from the Editorial Office was exceptional. From the initial submission to the final publication, I was guided through every step of the process with great care and professionalism. The team's responsiveness and assistance made the entire experience both easy and stress-free. I am also deeply impressed by the quality and reputation of the journal. It is an honor to have my research featured in such a respected publication, and I am confident that it will make a meaningful contribution to the field.

"I am grateful for the opportunity of contributing to [International Journal of Clinical Case Reports and Reviews] and for the rigorous review process that enhances the quality of research published in your esteemed journal. I sincerely appreciate the time and effort of your team who have dedicatedly helped me in improvising changes and modifying my manuscript. The insightful comments and constructive feedback provided have been invaluable in refining and strengthening my work".

I thank the ‘Journal of Clinical Research and Reports’ for accepting this article for publication. This is a rigorously peer reviewed journal which is on all major global scientific data bases. I note the review process was prompt, thorough and professionally critical. It gave us an insight into a number of important scientific/statistical issues. The review prompted us to review the relevant literature again and look at the limitations of the study. The peer reviewers were open, clear in the instructions and the editorial team was very prompt in their communication. This journal certainly publishes quality research articles. I would recommend the journal for any future publications.

Dear Jessica Magne, with gratitude for the joint work. Fast process of receiving and processing the submitted scientific materials in “Clinical Cardiology and Cardiovascular Interventions”. High level of competence of the editors with clear and correct recommendations and ideas for enriching the article.

We found the peer review process quick and positive in its input. The support from the editorial officer has been very agile, always with the intention of improving the article and taking into account our subsequent corrections.

My article, titled 'No Way Out of the Smartphone Epidemic Without Considering the Insights of Brain Research,' has been republished in the International Journal of Clinical Case Reports and Reviews. The review process was seamless and professional, with the editors being both friendly and supportive. I am deeply grateful for their efforts.
To Dear Erin Aust – Editorial Coordinator of Journal of General Medicine and Clinical Practice! I declare that I am absolutely satisfied with your work carried out with great competence in following the manuscript during the various stages from its receipt, during the revision process to the final acceptance for publication. Thank Prof. Elvira Farina

Dear Jessica, and the super professional team of the ‘Clinical Cardiology and Cardiovascular Interventions’ I am sincerely grateful to the coordinated work of the journal team for the no problem with the submission of my manuscript: “Cardiometabolic Disorders in A Pregnant Woman with Severe Preeclampsia on the Background of Morbid Obesity (Case Report).” The review process by 5 experts was fast, and the comments were professional, which made it more specific and academic, and the process of publication and presentation of the article was excellent. I recommend that my colleagues publish articles in this journal, and I am interested in further scientific cooperation. Sincerely and best wishes, Dr. Oleg Golyanovskiy.

Dear Ashley Rosa, Editorial Coordinator of the journal - Psychology and Mental Health Care. " The process of obtaining publication of my article in the Psychology and Mental Health Journal was positive in all areas. The peer review process resulted in a number of valuable comments, the editorial process was collaborative and timely, and the quality of this journal has been quickly noticed, resulting in alternative journals contacting me to publish with them." Warm regards, Susan Anne Smith, PhD. Australian Breastfeeding Association.

Dear Jessica Magne, Editorial Coordinator, Clinical Cardiology and Cardiovascular Interventions, Auctores Publishing LLC. I appreciate the journal (JCCI) editorial office support, the entire team leads were always ready to help, not only on technical front but also on thorough process. Also, I should thank dear reviewers’ attention to detail and creative approach to teach me and bring new insights by their comments. Surely, more discussions and introduction of other hemodynamic devices would provide better prevention and management of shock states. Your efforts and dedication in presenting educational materials in this journal are commendable. Best wishes from, Farahnaz Fallahian.
Dear Maria Emerson, Editorial Coordinator, International Journal of Clinical Case Reports and Reviews, Auctores Publishing LLC. I am delighted to have published our manuscript, "Acute Colonic Pseudo-Obstruction (ACPO): A rare but serious complication following caesarean section." I want to thank the editorial team, especially Maria Emerson, for their prompt review of the manuscript, quick responses to queries, and overall support. Yours sincerely Dr. Victor Olagundoye.

Dear Ashley Rosa, Editorial Coordinator, International Journal of Clinical Case Reports and Reviews. Many thanks for publishing this manuscript after I lost confidence the editors were most helpful, more than other journals Best wishes from, Susan Anne Smith, PhD. Australian Breastfeeding Association.

Dear Agrippa Hilda, Editorial Coordinator, Journal of Neuroscience and Neurological Surgery. The entire process including article submission, review, revision, and publication was extremely easy. The journal editor was prompt and helpful, and the reviewers contributed to the quality of the paper. Thank you so much! Eric Nussbaum, MD
Dr Hala Al Shaikh This is to acknowledge that the peer review process for the article ’ A Novel Gnrh1 Gene Mutation in Four Omani Male Siblings, Presentation and Management ’ sent to the International Journal of Clinical Case Reports and Reviews was quick and smooth. The editorial office was prompt with easy communication.

Dear Erin Aust, Editorial Coordinator, Journal of General Medicine and Clinical Practice. We are pleased to share our experience with the “Journal of General Medicine and Clinical Practice”, following the successful publication of our article. The peer review process was thorough and constructive, helping to improve the clarity and quality of the manuscript. We are especially thankful to Ms. Erin Aust, the Editorial Coordinator, for her prompt communication and continuous support throughout the process. Her professionalism ensured a smooth and efficient publication experience. The journal upholds high editorial standards, and we highly recommend it to fellow researchers seeking a credible platform for their work. Best wishes By, Dr. Rakhi Mishra.

Dear Jessica Magne, Editorial Coordinator, Clinical Cardiology and Cardiovascular Interventions, Auctores Publishing LLC. The peer review process of the journal of Clinical Cardiology and Cardiovascular Interventions was excellent and fast, as was the support of the editorial office and the quality of the journal. Kind regards Walter F. Riesen Prof. Dr. Dr. h.c. Walter F. Riesen.

Dear Ashley Rosa, Editorial Coordinator, International Journal of Clinical Case Reports and Reviews, Auctores Publishing LLC. Thank you for publishing our article, Exploring Clozapine's Efficacy in Managing Aggression: A Multiple Single-Case Study in Forensic Psychiatry in the international journal of clinical case reports and reviews. We found the peer review process very professional and efficient. The comments were constructive, and the whole process was efficient. On behalf of the co-authors, I would like to thank you for publishing this article. With regards, Dr. Jelle R. Lettinga.

Dear Clarissa Eric, Editorial Coordinator, Journal of Clinical Case Reports and Studies, I would like to express my deep admiration for the exceptional professionalism demonstrated by your journal. I am thoroughly impressed by the speed of the editorial process, the substantive and insightful reviews, and the meticulous preparation of the manuscript for publication. Additionally, I greatly appreciate the courteous and immediate responses from your editorial office to all my inquiries. Best Regards, Dariusz Ziora

Dear Chrystine Mejia, Editorial Coordinator, Journal of Neurodegeneration and Neurorehabilitation, Auctores Publishing LLC, We would like to thank the editorial team for the smooth and high-quality communication leading up to the publication of our article in the Journal of Neurodegeneration and Neurorehabilitation. The reviewers have extensive knowledge in the field, and their relevant questions helped to add value to our publication. Kind regards, Dr. Ravi Shrivastava.

Dear Clarissa Eric, Editorial Coordinator, Journal of Clinical Case Reports and Studies, Auctores Publishing LLC, USA Office: +1-(302)-520-2644. I would like to express my sincere appreciation for the efficient and professional handling of my case report by the ‘Journal of Clinical Case Reports and Studies’. The peer review process was not only fast but also highly constructive—the reviewers’ comments were clear, relevant, and greatly helped me improve the quality and clarity of my manuscript. I also received excellent support from the editorial office throughout the process. Communication was smooth and timely, and I felt well guided at every stage, from submission to publication. The overall quality and rigor of the journal are truly commendable. I am pleased to have published my work with Journal of Clinical Case Reports and Studies, and I look forward to future opportunities for collaboration. Sincerely, Aline Tollet, UCLouvain.

Dear Ms. Mayra Duenas, Editorial Coordinator, International Journal of Clinical Case Reports and Reviews. “The International Journal of Clinical Case Reports and Reviews represented the “ideal house” to share with the research community a first experience with the use of the Simeox device for speech rehabilitation. High scientific reputation and attractive website communication were first determinants for the selection of this Journal, and the following submission process exceeded expectations: fast but highly professional peer review, great support by the editorial office, elegant graphic layout. Exactly what a dynamic research team - also composed by allied professionals - needs!" From, Chiara Beccaluva, PT - Italy.

Dear Maria Emerson, Editorial Coordinator, we have deeply appreciated the professionalism demonstrated by the International Journal of Clinical Case Reports and Reviews. The reviewers have extensive knowledge of our field and have been very efficient and fast in supporting the process. I am really looking forward to further collaboration. Thanks. Best regards, Dr. Claudio Ligresti
Dear Chrystine Mejia, Editorial Coordinator, Journal of Neurodegeneration and Neurorehabilitation. “The peer review process was efficient and constructive, and the editorial office provided excellent communication and support throughout. The journal ensures scientific rigor and high editorial standards, while also offering a smooth and timely publication process. We sincerely appreciate the work of the editorial team in facilitating the dissemination of innovative approaches such as the Bonori Method.” Best regards, Dr. Matteo Bonori.

I recommend without hesitation submitting relevant papers on medical decision making to the International Journal of Clinical Case Reports and Reviews. I am very grateful to the editorial staff. Maria Emerson was a pleasure to communicate with. The time from submission to publication was an extremely short 3 weeks. The editorial staff submitted the paper to three reviewers. Two of the reviewers commented positively on the value of publishing the paper. The editorial staff quickly recognized the third reviewer’s comments as an unjust attempt to reject the paper. I revised the paper as recommended by the first two reviewers.

Dear Maria Emerson, Editorial Coordinator, Journal of Clinical Research and Reports. Thank you for publishing our case report: "Clinical Case of Effective Fetal Stem Cells Treatment in a Patient with Autism Spectrum Disorder" within the "Journal of Clinical Research and Reports" being submitted by the team of EmCell doctors from Kyiv, Ukraine. We much appreciate a professional and transparent peer-review process from Auctores. All research Doctors are so grateful to your Editorial Office and Auctores Publishing support! I amiably wish our article publication maintained a top quality of your International Scientific Journal. My best wishes for a prosperity of the Journal of Clinical Research and Reports. Hope our scientific relationship and cooperation will remain long lasting. Thank you very much indeed. Kind regards, Dr. Andriy Sinelnyk Cell Therapy Center EmCell

Dear Editorial Team, Clinical Cardiology and Cardiovascular Interventions. It was truly a rewarding experience to work with the journal “Clinical Cardiology and Cardiovascular Interventions”. The peer review process was insightful and encouraging, helping us refine our work to a higher standard. The editorial office offered exceptional support with prompt and thoughtful communication. I highly value the journal’s role in promoting scientific advancement and am honored to be part of it. Best regards, Meng-Jou Lee, MD, Department of Anesthesiology, National Taiwan University Hospital.

Dear Editorial Team, Journal-Clinical Cardiology and Cardiovascular Interventions, “Publishing my article with Clinical Cardiology and Cardiovascular Interventions has been a highly positive experience. The peer-review process was rigorous yet supportive, offering valuable feedback that strengthened my work. The editorial team demonstrated exceptional professionalism, prompt communication, and a genuine commitment to maintaining the highest scientific standards. I am very pleased with the publication quality and proud to be associated with such a reputable journal.” Warm regards, Dr. Mahmoud Kamal Moustafa Ahmed

Dear Maria Emerson, Editorial Coordinator of ‘International Journal of Clinical Case Reports and Reviews’, I appreciate the opportunity to publish my article with your journal. The editorial office provided clear communication during the submission and review process, and I found the overall experience professional and constructive. Best regards, Elena Salvatore.

Dear Mayra Duenas, Editorial Coordinator of ‘International Journal of Clinical Case Reports and Reviews Herewith I confirm an optimal peer review process and a great support of the editorial office of the present journal
Dear Editorial Team, Clinical Cardiology and Cardiovascular Interventions. I am really grateful for the peers review; their feedback gave me the opportunity to reflect on the message and impact of my work and to ameliorate the article. The editors did a great job in addition by encouraging me to continue with the process of publishing.

Dear Cecilia Lilly, Editorial Coordinator, Endocrinology and Disorders, Thank you so much for your quick response regarding reviewing and all process till publishing our manuscript entitled: Prevalence of Pre-Diabetes and its Associated Risk Factors Among Nile College Students, Sudan. Best regards, Dr Mamoun Magzoub.

International Journal of Clinical Case Reports and Reviews is a high quality journal that has a clear and concise submission process. The peer review process was comprehensive and constructive. Support from the editorial office was excellent, since the administrative staff were responsive. The journal provides a fast and timely publication timeline.

Dear Maria Emerson, Editorial Coordinator of International Journal of Clinical Case Reports and Reviews, What distinguishes International Journal of Clinical Case Report and Review is not only the scientific rigor of its publications, but the intellectual climate in which research is evaluated. The submission process is refreshingly free of unnecessary formal barriers and bureaucratic rituals that often complicate academic publishing without adding real value. The peer-review system is demanding yet constructive, guided by genuine scientific dialogue rather than hierarchical or authoritarian attitudes. Reviewers act as collaborators in improving the manuscript, not as gatekeepers imposing arbitrary standards. This journal offers a rare balance: high methodological standards combined with a respectful, transparent, and supportive editorial approach. In an era where publishing can feel more burdensome than research itself, this platform restores the original purpose of peer review — to refine ideas, not to obstruct them Prof. Perlat Kapisyzi, FCCP PULMONOLOGIST AND THORACIC IMAGING.

Dear Grace Pierce, International Journal of Clinical Case Reports and Reviews I appreciate the opportunity to review for Auctore Journal, as the overall editorial process was smooth, transparent and professionally managed. This journal maintains high scientific standards and ensures timely communications with authors, which is truly commendable. I would like to express my special thanks to editor Grace Pierce for his constant guidance, promt responses, and supportive coordination throughout the review process. I am also greatful to Eleanor Bailey from the finance department for her clear communication and efficient handling of all administrative matters. Overall, my experience with Auctore Journal has been highly positive and rewarding. Best regards, Sabita sinha

Dear Mayra Duenas, Editorial Coordinator of the journal IJCCR, I write here a little on my experience as an author submitting to the International Journal of Clinical Case Reports and Reviews (IJCCR). This was my first submission to IJCCR and my manuscript was inherently an outsider’s effort. It attempted to broadly identify and then make some sense of life’s under-appreciated mysteries. I initially had responded to a request for possible submissions. I then contacted IJCCR with a tentative topic for a manuscript. They quickly got back with an approval for the submission, but with a particular requirement that it be medically relevant. I then put together a manuscript and submitted it. After the usual back-and-forth over forms and formality, the manuscript was sent off for reviews. Within 2 weeks I got back 4 reviews which were both helpful and also surprising. Surprising in that the topic was somewhat foreign to medical literature. My subsequent updates in response to the reviewer comments went smoothly and in short order I had a series of proofs to evaluate. All in all, the whole publication process seemed outstanding. It was both helpful in terms of the paper’s content and also in terms of its efficient and friendly communications. Thank you all very much. Sincerely, Ted Christopher, Rochester, NY.
